BALs are prognostic biomarkers and correlate with malignant behaviors in breast cancer
- PMID: 40707929
- PMCID: PMC12288338
- DOI: 10.1186/s12885-025-14576-0
BALs are prognostic biomarkers and correlate with malignant behaviors in breast cancer
Abstract
Background: The B-aggressive lymphoma (BAL) proteins, including BAL1, BAL2, and BAL3, constitute a conserved protein family characterized by their N-terminal macro domains and putative C-terminal poly (ADP-ribose) polymerase (PARP) active site. Dysregulation of BALs has been closely associated with the progression of various cancers. However, there is limited understanding of their precise expression profile, prognostic significance, and role in breast cancer (BC).
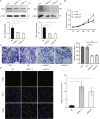
Methods: The expression patterns of BALs were evaluated utilizing multiple databases, including Ualcan, Gene Set Cancer Analysis (GSCA), Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), and Gene Expression Profiling Interactive Analysis (GEPIA). The prognostic significance of BALs was assessed via Kaplan-Meier plotter analysis. Furthermore, the potential mechanisms underlying the contribution of BC progression were predicted through GO and KEGG pathway enrichment analysis. Additionally, the effect of BALs on the malignant behaviors of BC cells was determined using CCK-8 assay, Transwell assay, and TUNEL assay.
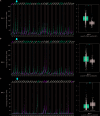
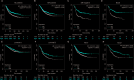
Results: The data revealed that the expression levels of both BAL1 and BAL2 were upregulated in BC, whereas no significant change was observed for BAL3. Survival analysis demonstrated a strong association between the overexpression of both BAL1 and BAL2 and favorable prognosis in patients with various subtypes of BC, including estrogen receptor (ER)-positive, ER-negative, Basal, luminal B, HER2-, and HER2 + subtypes. Additionally, the knockdown of BAL1 and BAL2 inhibited the proliferation and migration of BC cells while facilitating apoptosis.
Conclusions: These findings suggest that both BAL1 and BAL2 hold great potential as significant prognostic biomarkers and therapeutic targets for patients with BC.
Keywords: B-aggressive lymphoma protein; Biomarker; Breast cancer; Prognosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study does not involve animal and human ethics. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2024;74(3):229–63. - PubMed
-
- Liu Y, Li Y, Du C, Kuang S, Zhou X, Zhang J, et al. Underlying mechanisms of epithelial splicing regulatory proteins in cancer progression. J Mol Med (Berl). 2022;100(11):1539–56. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

