Leveraging unmanned aerial vehicle derived multispectral data for improved genomic prediction in potato (Solanum tuberosum)
- PMID: 40708258
- PMCID: PMC12290324
- DOI: 10.1002/tpg2.70082
Leveraging unmanned aerial vehicle derived multispectral data for improved genomic prediction in potato (Solanum tuberosum)
Abstract
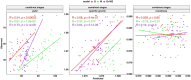
Multispectral leaf canopy reflectance as measured by unmanned aerial vehicles is the result of genetic and environmental interactions driving plant physiochemical processes. These measures can then be used to construct relationship matrices for modeling genetic main effects. This type of phenotypic prediction is particularly relevant for trials with many entries, such as those used in early generation potato (Solanum tuberosum) breeding. We compared three methods for making predictions in our potato breeding program: first, using multispectral-derived relationship matrices; second, using the traditional approach based on genomic derived relationships; and third, using a combination of both. Multispectral bands were collected at five different time points for two market classes of potato: chipping and fresh market. We modeled genetic main effects for yield and quality traits at each time point and all stages combined. Models with multispectral relationship matrices exhibited better prediction accuracy for yield and roundness than genomic only models and models featuring spectra plus genomic kernels outperformed both single-kernel predictions in terms of accuracy for most traits. Time points were variably informative depending on the trait measured, however, for all traits combining across time points performed as well or better than single time point models. Similarly, using feature selection to limit our models to important variables did not improve prediction accuracy significantly. This work highlights two potential uses for spectral data in genomic prediction: first, as an alternative to genetic data and second, in combination with genetic data to increase precision of selection.
© 2025 The Author(s). The Plant Genome published by Wiley Periodicals LLC on behalf of Crop Science Society of America.
Conflict of interest statement
Laura M. Shannon is a member of the editorial board of The Plant Genome.
Figures






References
-
- Abdelhakim, L. O. A. , Pleskačová, B. , Rodriguez‐Granados, N. Y. , Sasidharan, R. , Perez‐Borroto, L. S. , Sonnewald, S. , Gruden, K. , Vothknecht, U. C. , Teige, M. , & Panzarová, K. (2024). High throughput image‐based phenotyping for determining morphological and physiological responses to single and combined stresses in potato. Journal of Visualized Experiments, 208, e66255. 10.3791/66255 - DOI - PubMed
-
- Agha, H. I. , Endelman, J. B. , Chitwood‐Brown, J. , Clough, M. , Coombs, J. , De Jong, W. S. , Douches, D. S. , Higgins, C. R. , Holm, D. G. , Novy, R. , Resende, M. F. R. , Sathuvalli, V. , Thompson, A. L. , Yencho, G. C. , Zotarelli, L. , & Shannon, L. M. (2024). Genotype‐by‐environment interactions and local adaptation shape selection in the US National Chip Processing Trial. Theoretical and Applied Genetics [Theoretische Und Angewandte Genetik], 137(5), Article 99. 10.1007/s00122-024-04610-3 - DOI - PMC - PubMed
-
- Alemu, A. , Åstrand, J. , Montesinos‐López, O. A. , Isidro Y Sánchez, J. , Fernández‐Gónzalez, J. , Tadesse, W. , Vetukuri, R. R. , Carlsson, A. S. , Ceplitis, A. , Crossa, J. , Ortiz, R. , & Chawade, A. (2024). Genomic selection in plant breeding: Key factors shaping two decades of progress. Molecular Plant, 17(4), 552–578. 10.1016/j.molp.2024.03.007 - DOI - PubMed
-
- Alkhaled, A. , Townsend, P. A. , & Wang, Y. (2023). Remote sensing for monitoring potato nitrogen status. American Journal of Potato Research, 100(1), 1–14. 10.1007/s12230-022-09898-9 - DOI
-
- Asano, K. , & Endelman, J. B. (2023). Development of KASP markers for the potato virus Y resistance gene Rychc using whole‐genome resequencing data. bioRxiv. 10.1101/2023.12.20.572658 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

