Diversity and functional roles of endophytic and rhizospheric microorganisms in Ophioglossum vulgatum L.: implications for bioactive compound synthesis
- PMID: 40708922
- PMCID: PMC12288683
- DOI: 10.3389/fmicb.2025.1618667
Diversity and functional roles of endophytic and rhizospheric microorganisms in Ophioglossum vulgatum L.: implications for bioactive compound synthesis
Abstract
Background: Ophioglossum vulgatum L. is a widely utilized medicinal plant, with the entire plant being used for medicinal purposes. This study systematically characterized the endophytic and rhizospheric community structure, taxonomic diversity, and symbiotic networks within distinct compartments of O. vulgatum, while evaluating their potential associations with the accumulation of pharmacologically active metabolites.
Methods: Endophytic and rhizospheric community profiling was conducted via Illumina sequencing, while bioactive compounds were identified using UPLC-ESI-MS/MS.
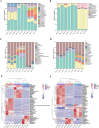
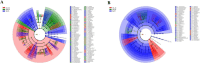
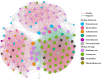
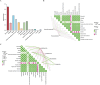
Results: Roots and leaves harbored beneficial bacteria (e.g., Methylobacterium, Streptomyces, Sphingomonas, and Flavobacterium). Dominant fungi included Archaeorhizomyces (rhizosphere soil) and Homophron (roots/leaves). Dark septate endophytes (DSEs; e.g., Cladosporium, Cladophialophora, and Chaetomium) were abundant across rhizosphere soil, roots, and leaves. Alpha/beta diversity analyses showed higher microbial richness in rhizosphere soil than in plant tissues. Functional predictions (PICRUSt2/FUNGuild) linked endophytic and rhizospheric bacteria to metabolism, human diseases, and biological systems. Network analysis highlighted Basidiomycota as keystone taxa, with modular community structure. Functional predictions revealed that endophytic and rhizospheric microorganisms were associated with critical metabolic pathways, particularly in the biosynthesis of flavonoids and alkaloids (primary bioactive compounds). LEFSe analyses highlighted compartment-specific biomarkers: Acidobacteria, Basidiomycota, and Ascomycota were enriched in distinct zones (rhizosphere, roots, and leaves), with Actinobacteria exhibiting highly significant correlations (P < 0.01) with flavonoids, lipids, and quinones, while Acidobacteria, Basidiomycota, and Ascomycota were strongly linked to steroids and tannins (P < 0.05).
Conclusion: The diversity and abundance of microbial communities in O. vulgatum exhibited tissue-specific and rhizosphere-dependent variations, with distinct patterns strongly correlating to bioactive compound accumulation. Notably, biomarker taxa including Actinobacteria, Acidobacteria, Basidiomycota, and Ascomycota demonstrated robust microbe-metabolite interactions, suggesting their critical regulatory role in biosynthesis pathways. These findings establish endophytic-rhizospheric microbiota as key biosynthetic modulators, proposing innovative approaches for enhancing phytochemical production through targeted microbial community manipulation.
Keywords: Ophioglossum vulgatum; bioactive compounds; community diversity; plant endophytes; rhizosphere soil.
Copyright © 2025 Long, Zhang, Wu, Tang, Zheng, Chen, Cao, Zhou and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abiven S., Menasseri S., Angers D. A., Leterme P. (2007). Dynamics of aggregate stability and biological binding agents during decomposition of organic materials. Eur. J. Soil Sci. 58 239–247. 10.1111/j.1365-2389.2006.00833.x - DOI
LinkOut - more resources
Full Text Sources

