Association analysis of single nucleotide polymorphisms in the LIFR gene with lambing number in sheep
- PMID: 40708997
- PMCID: PMC12287739
- DOI: 10.3389/fvets.2025.1629162
Association analysis of single nucleotide polymorphisms in the LIFR gene with lambing number in sheep
Abstract
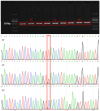
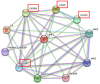
Lambing number trait in sheep is a complex trait controlled by multiple genes, and low lambing number has a severe economic impact on the sheep industry. Previous studies have shown that Sparc/Osteonectin, cwcv, and kazal-like domain proteoglycan 1 (SPOCK1), A disintegrin and metalloproteinase with thrombospondin-like repeats 1 (ADAMTS1), heparin-binding epidermal growth factor-like growth factor (HBEGF), and leukemia inhibitory factor receptor (LIFR) are involved in mammalian reproduction. However, the effects of these genes on lambing number in sheep are still unclear. In this study, single nucleotide polymorphism (SNP) loci at the above four genes were genotyped in five sheep breeds (two single-born sheep breeds and three multiple-born sheep breeds with a total of 768 sheep) using the Sequenom MassARRAY® SNP assay, and their associations with the lambing number in small-tailed Han sheep were analyzed. The results showed that a total of six SNP loci (c.*1633A>G, c.*1388T>C, c.*1095G>A, c.1847A>G, c.*2403A>G, and c.*127T>C) were identified in the four genes, SPOCK1, ADAMTS1, HBEGF, and LIFR. All the above SNPs had three genotypes in five sheep breeds. The population genetic analysis of SNPs of the four genes and the association analysis with lambing number showed that the polymorphism information content of the five sheep breeds of small-tailed Han sheep, Hu sheep, Cele black sheep, Sunite sheep, and Bamei mutton sheep was between 0 and 0.37, and a few sheep breeds were in Hardy-Weinberg equilibrium (p>0.05). The c.*127 T>C locus of the LIFR gene may be affected by natural or artificial selection in these sheep breeds. Additionally, the association analysis between the c.*127 T>C locus of LIFR gene and the lambing number of small-tailed Han sheep showed that the c.*127 T>C locus of the LIFR gene was significantly associated with the lambing number of the second, third, and average parity of smalltailed Han sheep (p < 0.05). The lambing number of ewes with the LIFR CC genotype was significantly lower than that with the TT and CT genotypes (p < 0.05). The LIFR protein interaction network was also predicted and found to interact with the reported ciliary neurotrophic factor (CNTF), cardiotrophinlike cytokine factor 1 (CLCF1), interleukin 6 cytokine family signal transducer (IL6ST), and ciliary neurotrophic factor receptor (CNTFR) proteins. In conclusion, the c.*127 T>C locus of LIFR gene can be used as a candidate genetic molecular marker to increase lambing numbers in polytocous sheep.
Keywords: LIFR; MassARRAY; lambing number; molecular marker; sheep.
Copyright © 2025 Wen, Xiang, Zhang, Liu, Liu and Chu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

