Temporal TCR dynamics and epitope diversity mark recovery in severe COVID-19 patients
- PMID: 40709175
- PMCID: PMC12286809
- DOI: 10.3389/fimmu.2025.1582949
Temporal TCR dynamics and epitope diversity mark recovery in severe COVID-19 patients
Abstract
Introduction: Severe COVID-19 is characterized by immune dysregulation, with T cells playing a central role in disease progression and recovery. However, the longitudinal dynamics of the T cell receptor (TCR) repertoire during the course of severe illness remain unclear.
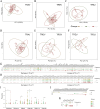
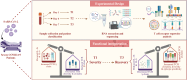
Methods: To investigate temporal changes in adaptive immunity, we analyzed peripheral blood samples from the ICU-admitted severe COVID-19 patients (n = 36) collected at three time points: Day 1 (T1), Day 4 (T2), and Day 7 (T3). Bulk RNA-sequencing was performed to extract TCR repertoires, and cytokine profiles were assessed in parallel. TCR clonotypes were annotated using VDJdb and TCRex to infer potential epitope specificities.
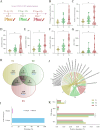
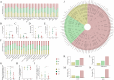
Results: By T3, we observed a 2.3-fold expansion in TCR clonotypes along with increased TCR-β (TRB) chain usage, indicating the emergence of a broad polyclonal T cell response. In contrast, TCR-γ (TRG) chain prevalence declined. Pro-inflammatory cytokines, including IL-1β and IL-6, were reduced over time, marking a shift toward immune resolution. Changes in CDR3 motifs and preferential TRBV gene segment usage were detected, suggesting repertoire adaptation. Additionally, annotated TCR clonotypes at T3 mapped to SARS-CoV-2 and other pathogen-associated epitopes (e.g., CMV, Plasmodium), reflecting possible cross-reactivity or memory T cell recruitment.
Discussion: These findings suggest a coordinated transition from immune dysfunction to recovery in severe COVID-19, marked by expanding TCR diversity, reduced inflammation, and predicted broadening of antigen recognition. The integrated analysis of TCR repertoire dynamics and cytokine profiles provides insights into the adaptive immune mechanisms underlying viral clearance and immune stabilization.
Keywords: COVID-19; ICU-admitted patients; TCR clonotypes; TCR dynamics; adaptive regulation.
Copyright © 2025 Khare, Yadav, Halder, Ray, Ganguly and Pandey.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
High TCR Degeneracy Enhances Antiviral Efficacy of HTLV-1-Specific CTLs by Targeting Variant Viruses in HAM Patients.Int J Mol Sci. 2025 Jul 10;26(14):6602. doi: 10.3390/ijms26146602. Int J Mol Sci. 2025. PMID: 40724852 Free PMC article.
-
Nonconserved epitopes dominate reverse preexisting T cell immunity in COVID-19 convalescents.Signal Transduct Target Ther. 2024 Jun 12;9(1):160. doi: 10.1038/s41392-024-01876-3. Signal Transduct Target Ther. 2024. PMID: 38866784 Free PMC article.
-
Impact of CD4+ T cell and TCR repertoires on SARS-CoV-2-Specific antibody responses in PLWH following COVID-19 vaccination.J Immunol. 2025 May 1;214(5):917-925. doi: 10.1093/jimmun/vkae040. J Immunol. 2025. PMID: 40235093
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Accuracy of routine laboratory tests to predict mortality and deterioration to severe or critical COVID-19 in people with SARS-CoV-2.Cochrane Database Syst Rev. 2024 Aug 6;8(8):CD015050. doi: 10.1002/14651858.CD015050.pub2. Cochrane Database Syst Rev. 2024. PMID: 39105481 Free PMC article.
References
-
- Carter C, Notter J. COVID-19 Disease: a critical care perspective. Clinics Integrated Care. (2020) 1:100003. doi: 10.1016/j.intcar.2020.100003 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

