Comprehensive analysis of phosducin-like 3 as a diagnostic, prognostic and immunological marker in pan-cancer
- PMID: 40709189
- PMCID: PMC12287076
- DOI: 10.3389/fimmu.2025.1604179
Comprehensive analysis of phosducin-like 3 as a diagnostic, prognostic and immunological marker in pan-cancer
Abstract
Background: Phosducin-like 3 (PDCL3), a member of the photoreceptor family, is involved in angiogenesis and apoptosis. However, there is no pan-cancer analysis, and few studies have explored the effect of PDCL3 on tumor immune infiltration.
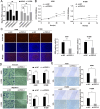
Method: Public datasets were used to explore the diagnostic and prognostic value of PDCL3. The relationship between PDCL3 expression and immune infiltration, tumor mutation burden (TMB), and microsatellite instability (MSI) was investigated. Additionally, the therapeutic value of PDCL3 was explored. Finally, differences in PDCL3 expression across cell clusters were analyzed using single-cell datasets. In vitro cellular assays were performed to assess the impact of PDCL3 expression on the proliferative capacity, migratory potential, and invasive properties of non-small cell lung cancer (NSCLC) cells.
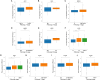
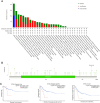
Results: PDCL3 expression was upregulated in most tumors and correlated with poor outcomes, showing diagnostic and prognostic value. In addition, PDCL3 expression exhibited a positive correlation with infiltration of T helper 2 (Th2) cells and a negative correlation with infiltration of plasmacytoid dendritic cells (pDCs) across a variety of tumors. A relationship was also found between PDCL3 expression and TMB and MSI. Single-cell dataset analysis confirmed that PDCL3 expression was primarily in cancer cells and macrophages. In vitro functional analyses demonstrated that genetic silencing of PDCL3 significantly reduced proliferative rates, migratory activity, and invasive potential in pulmonary carcinoma cell models.
Conclusions: PDCL3 may contribute to cancer progression and is a potential candidate biomarker for pan-cancer diagnosis and prognosis. These findings suggest that targeting PDCL3 may provide a valuable strategy for cancer immunotherapy.
Keywords: PDCL3; bioinformatics analysis; biomarker; immune infiltration; pan-cancer.
Copyright © 2025 Li, Li, Li, Liang, Wu, Zhu, Nong, Zhuo, Luo, He, Huang and Cao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

