This is a preprint.
Associations of proteomic age with mortality and incident chronic diseases in the European Prospective Investigation into Cancer and Nutrition (EPIC)
- PMID: 40709259
- PMCID: PMC12288520
- DOI: 10.21203/rs.3.rs-7087230/v1
Associations of proteomic age with mortality and incident chronic diseases in the European Prospective Investigation into Cancer and Nutrition (EPIC)
Abstract
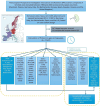
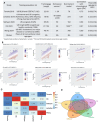
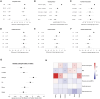
Assessment of biological ageing using proteomic clocks may enhance risk prediction and elucidate the molecular links between ageing and chronic diseases. Within a pre-diagnostic cohort of 17,473 Europeans with up to 28 years of follow-up, we examined associations of plasma SomaScan-based proteomic clocks, including organ-specific clocks, with 24 incident chronic diseases, all-cause mortality, and lifestyle risk factors. Global proteomic age gap (a composite biological age acceleration score combining previously published clocks) showed the strongest positive association of all tested clocks with all-cause mortality. Accelerated proteomic ageing was significantly associated with smoking, alcohol consumption, physical inactivity, and higher risk of cardiovascular diseases, dementia, and liver, upper aero-digestive tract, lung, and kidney cancers. Some organ-specific cancers were more strongly associated with their respective organ-specific age gaps. Mortality prediction by proteomic clocks was comparable in performance to classical lifestyle risk factors. In summary, proteomic clocks appear promising biomarkers of generalized age-related disease risk.
Keywords: Aging; SomaLogic; aptamers; biological age; biological clocks; cancer; cardiovascular disease; diabetes; neurodegeneration; proteomics; risk factors; risk prediction.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

