Epigenetic Regulation of B Cell Memory Formation: A Poised Model for B Cell Epigenetic Reprograming
- PMID: 40709286
- PMCID: PMC12289371
- DOI: 10.1155/jimr/9328523
Epigenetic Regulation of B Cell Memory Formation: A Poised Model for B Cell Epigenetic Reprograming
Abstract
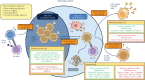
The formation of B cell immunological memory happens after the first encounter with a pathogen. At the germinal center (GC), B cells experience complex transcriptional and epigenetic transitions to differentiate into memory B cells (MBCs) and plasma cells (PCs). In particular, the differentiation of GC B cells into MBCs has been poorly understood, and no clear conclusions on the signals and transcription factors leading to this cell fate have been identified. Recent discoveries in epigenetics and immune memory have elucidated the essential role of epigenetic regulators in establishing the memory B cell (MBC) fate. DNA methylation regulators, histone modifiers, noncoding RNAs (ncRNAs), and chromatin remodelers orchestrate a dynamic reprograming of the MBC phenotype. Positive and negative epigenetic regulators of the B cell program collaborate at each differentiation stage and allow for complex chromatin topology rearrangements and dynamic exposure to transcription and translation. Following MBC fate determination at the GC, the acquired epigenetic modifications induce a poised regulatory state where genes are epigenetically marked to remain transcriptionally inactive, but primed for rapid activation upon stimuli. Thus, a poised epigenetic control over gene expression governs MBC formation and a novel model of epigenetic reprograming is proposed. This model provides a novel perspective on how the B cell fate is determined in the GC and memory is formed, offering insights for improved vaccination and therapeutical approaches.
Keywords: B cell fate determination; B cell memory; epigenetics; germinal center; memory B cells; poised genes.
Copyright © 2025 Carlos Valverde-Hernandez. Journal of Immunology Research published by John Wiley & Sons Ltd.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
Similar articles
-
Stage-specific DNA methylation dynamics in mammalian heart development.Epigenomics. 2025 Apr;17(5):359-371. doi: 10.1080/17501911.2025.2467024. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39980349 Review.
-
EZH2 coordinates memory B-cell programming and recall responses.J Immunol. 2025 May 1;214(5):947-957. doi: 10.1093/jimmun/vkaf004. J Immunol. 2025. PMID: 40073167
-
Bath: a Bayesian approach to analyze epigenetic transitions reveals a dual role of H3K27me3 in chondrogenesis.Epigenetics Chromatin. 2025 Jun 27;18(1):38. doi: 10.1186/s13072-025-00594-6. Epigenetics Chromatin. 2025. PMID: 40571950 Free PMC article.
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
Conserved Epigenetic Programming and Enhanced Heme Metabolism Drive Memory B Cell Reactivation.J Immunol. 2021 Apr 1;206(7):1493-1504. doi: 10.4049/jimmunol.2000551. Epub 2021 Feb 24. J Immunol. 2021. PMID: 33627377 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

