Clonal hematopoiesis in AML long-term survivors: Risk factors and clinical consequences
- PMID: 40709316
- PMCID: PMC12288099
- DOI: 10.1002/hem3.70183
Clonal hematopoiesis in AML long-term survivors: Risk factors and clinical consequences
Abstract
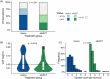
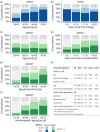
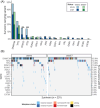
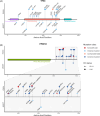
Clonal hematopoiesis (CH) is common in the general population and associated with various health risks, but its prevalence and clinical implications in acute myeloid leukemia (AML) long-term survivors (LTS; ≥5-year survival) are unknown. We analyzed CH in 373 AML-LTS with a median 11.6-year follow-up from diagnosis using a sensitive targeted sequencing assay based on single-molecule molecular inversion probes. CH variants were detected in 61.9% of survivors, with 26% having small-clone CH (SC-CH, variant allele frequency (VAF) < 2%) and 35.9% CH of indeterminate potential (≥2% VAF). CH was more prevalent in survivors treated with chemotherapy only (75.7%) compared to those who received allogeneic stem cell transplantation (alloSCT, 54.0%) and to age group-matched healthy controls. In chemotherapy-treated survivors, CH prevalence increased with age, whereas in alloSCT recipients, it most closely associated with hematopoietic age (i.e., the sum of donor age and time since transplantation). The variant spectrum also differed by treatment, with TP53 and PPM1D variants being more common in the chemotherapy group. CH variants ≥10% VAF associated with increased risks of diabetes in alloSCT recipients and secondary neoplasms in chemotherapy-treated survivors. This study provides insights into the high prevalence and potential clinical relevance of CH in AML-LTS.
© 2025 The Author(s). HemaSphere published by John Wiley & Sons Ltd on behalf of European Hematology Association.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
