Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring
- PMID: 40710080
- PMCID: PMC12293380
- DOI: 10.3390/bios15070430
Protein, Nucleic Acid, and Nanomaterial Engineering for Biosensors and Monitoring
Abstract
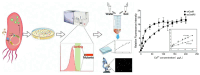
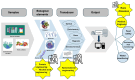
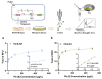
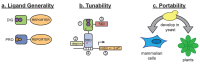
The engineering of proteins, nucleic acids, and nanomaterials has significantly advanced the development of biosensors for the monitoring of rare diseases. These innovative biosensing technologies facilitate the early detection and management of conditions that often lack adequate diagnostic solutions. By utilizing engineered proteins and functional nucleic acids, such as aptamers and nucleic acid sensors, these biosensors can achieve high specificity in identifying the biomarkers associated with rare diseases. The incorporation of nanomaterials, like nanoparticles and nanosensors, enhances sensitivity and allows for the real-time monitoring of biochemical changes, which is critical for timely intervention. Moreover, integrating these technologies into wearable devices provides patients and healthcare providers with continuous monitoring capabilities, transforming the landscape of healthcare for rare diseases. The ability to detect low-abundance biomarkers in varied sample types, such as blood or saliva, can lead to breakthroughs in understanding disease pathways and personalizing treatment strategies. As the field continues to evolve, the combination of protein, nucleic acid, and nanomaterial engineering will play a crucial role in developing next-generation biosensors that are not only cost-effective but also easy to use, ultimately improving outcomes and the quality of life for individuals affected by rare diseases.
Keywords: DNA; MOF; RNA; biosensor; directed evolution; nanocomposites; rare diseases.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Nucleic Acid Nanocapsules as a New Platform to Deliver Therapeutic Nucleic Acids for Gene Regulation.Acc Chem Res. 2025 Jul 1;58(13):1951-1962. doi: 10.1021/acs.accounts.5c00126. Epub 2025 Jun 9. Acc Chem Res. 2025. PMID: 40491030
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Nucleic acid-based wearable and implantable electrochemical sensors.Chem Soc Rev. 2024 Jul 29;53(15):7960-7982. doi: 10.1039/d4cs00001c. Chem Soc Rev. 2024. PMID: 38985007 Free PMC article. Review.
References
-
- Shen L., Chen Y.W., Pan J.J., Yu X., Zhang Y.B., Guo B.X., Wang J.Q., Liu Y., Xiao X., Chen S.P., et al. Development of a highly sensitive PbrR-based biosensor via directed evolution and its application for lead detection. J. Hazard. Mater. 2025;488:137489. doi: 10.1016/j.jhazmat.2025.137489. - DOI - PubMed
-
- Paulmurugan R., Afjei R., Sekar T.V., Babikir H.A., Massoud T.F. A protein folding molecular imaging biosensor monitors the effects of drugs that restore mutant p53 structure and its downstream function in glioblastoma cells. Oncotarget. 2018;9:21495–21511. doi: 10.18632/oncotarget.25138. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

