Genome Mining Reveals a Sactipeptide Biosynthetic Cluster in Staphylococcus pseudintermedius
- PMID: 40711295
- PMCID: PMC12300986
- DOI: 10.3390/vetsci12070635
Genome Mining Reveals a Sactipeptide Biosynthetic Cluster in Staphylococcus pseudintermedius
Abstract
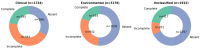
Staphylococcus pseudintermedius, an opportunistic pathogen of veterinary and zoonotic concern, harbors diverse biosynthetic gene clusters (BGCs) that may contribute to its ecological fitness and virulence. In this study, we performed a comparative genomic analysis of 6815 S. pseudintermedius isolates. Using Roary, we identified core and accessory genomes, revealing the subtilosin A gene (sboA) as part of the accessory genome, present in a subset of S. pseudintermedius isolates from clinical (n = 657), environmental (n = 1031), and unclassified sources (n = 487). AntiSMASH v8.0.0 analysis confirmed the presence of subtilosin A BGCs annotated as a sactipeptide with low similarity confidence to Bacillus subtilis subsp. spizizenii ATCC 6633 subtilosin A cluster. Further characterization using BAGEL4 identified multiple genes homologous to the Bacillus subtilis subtilosin A biosynthetic machinery (sbo-albABCDEFG), although albB, albG, and sboX were not annotated, raising questions about cluster completeness and functionality. BLAST v2.12.0 analysis of the full BGC identified by BAGEL4, revealing high conservation (99.6-100% pairwise identity) of gene content and order in 395 clinical, 593 environmental, and 417 unclassified S. pseudintermedius isolates. Incomplete clusters were identified in 763 clinical, 942 environmental, and 201 unclassified S. pseudintermedius isolates. The discrepancy between the number of isolates containing sboA and those harboring the full cluster may reflect evolutionary divergence or could be attributed to limitations in assembly quality. The functional implications of the identified cluster in S. pseudintermedius remain to be elucidated; however, its potential role in conferring competitive fitness by inhibiting closely related species is supported by previous findings in other staphylococci.
Keywords: Staphylococcus pseudintermedius; antibiotics resistance; bacteriocin; subtilosin A.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Silva V., Oliveira A., Manageiro V., Caniça M., Contente D., Capita R., Alonso-Calleja C., Carvalho I., Capelo J.L., Igrejas G., et al. Clonal Diversity and Antimicrobial Resistance of Methicillin-Resistant Staphylococcus pseudintermedius Isolated from Canine Pyoderma. Microorganisms. 2021;9:482. doi: 10.3390/microorganisms9030482. - DOI - PMC - PubMed
-
- Viñes J., Verdejo M.Á., Horvath L., Vergara A., Vila J., Francino O., Morata L., Espasa M., Casals-Pascual C., Soriano À., et al. Isolation of Staphylococcus pseudintermedius in Immunocompromised Patients from a Single Center in Spain: A Zoonotic Pathogen from Companion Animals. Microorganisms. 2024;12:1695. doi: 10.3390/microorganisms12081695. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

