Isolation and Biological Characteristics Study of Porcine Reproductive and Respiratory Syndrome Virus GZ2022 Strain
- PMID: 40711311
- PMCID: PMC12299216
- DOI: 10.3390/vetsci12070651
Isolation and Biological Characteristics Study of Porcine Reproductive and Respiratory Syndrome Virus GZ2022 Strain
Abstract
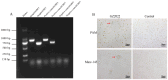
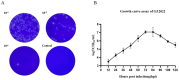
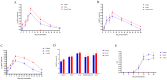
PRRSV continues to evolve, complicating its epidemiological landscape in China. In this study, we isolated a novel PRRSV strain, GZ2022, from a swine farm in Guizhou Province. Subsequent analyses performed on this isolate included complete genome sequencing, phylogenetic analysis, recombination assessment, and characterization of its biological properties. Phylogenetic analysis revealed that GZ2022 clusters within Lineage 1 (NADC30-like) and features a 131-amino-acid deletion in NSP2, consistent with NADC30-derived strains. Recombination analysis identified NADC30 as the major parental strain (75% genomic contribution), with a minor recombinant region (25%) derived from the highly pathogenic HuN4 strain. In vitro growth kinetics revealed peak viral titers in Marc-145 cells at 72 h post infection (hpi). Pathogenicity was evaluated in 21-day-old piglets infected with GZ2022, the highly pathogenic PRRSV strain WUH3, or negative controls. Both infected groups exhibited typical PRRS clinical signs (fever, respiratory distress) and histopathological lesions (interstitial pneumonia, pulmonary consolidation). However, GZ2022-infected piglets exhibited attenuated virulence compared to WUH3, with reduced pulmonary hemorrhage and 0% mortality compared to 80% in the WUH3 group. Seroconversion (N-protein antibodies) was observed at 14 dpi (days post inoculation) in GZ2022-infected animals, persisting throughout the 28-day trial. Viral shedding dynamics aligned with moderate pathogenicity. These findings classify GZ2022 as a moderately virulent NADC30-like recombinant strain with partial HuN4-derived genomic regions. The emergence of such strains underscores the need for sustained surveillance of PRRSV genetic diversity and systematic evaluation of the biological properties of novel variants to refine control measures and inform vaccine development.
Keywords: GZ2022; PRRSV; complete genome sequencing; genetic variation; pathogenicity; phylogenetic analysis.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures






Similar articles
-
A novel L1C.5 RFLP-1-4-4 recombinant porcine reproductive and respiratory syndrome virus between wild-type virus and a modified-live virus vaccine is highly pathogenic to piglets.Front Vet Sci. 2025 Jul 2;12:1627238. doi: 10.3389/fvets.2025.1627238. eCollection 2025. Front Vet Sci. 2025. PMID: 40671821 Free PMC article.
-
Etiological Detection, Isolation, and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus in China.Vet Sci. 2025 May 29;12(6):530. doi: 10.3390/vetsci12060530. Vet Sci. 2025. PMID: 40559767 Free PMC article.
-
Genome and Pathogenicity Analysis of an NADC30-like PRRSV Strain in China's Xinjiang Province.Viruses. 2025 Mar 6;17(3):379. doi: 10.3390/v17030379. Viruses. 2025. PMID: 40143307 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Li J., Wang S., Li C., Wang C., Liu Y., Wang G., He X., Hu L., Liu Y., Cui M., et al. Secondary Haemophilus parasuis infection enhances highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) infection-mediated inflammatory responses. Vet. Microbiol. 2017;4:35–42. doi: 10.1016/j.vetmic.2017.03.035. - DOI - PubMed
LinkOut - more resources
Full Text Sources

