Omicron Impacts Olfaction in Hamsters
- PMID: 40711352
- PMCID: PMC12292033
- DOI: 10.1096/fj.202501431RR
Omicron Impacts Olfaction in Hamsters
Abstract
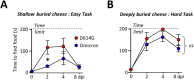
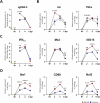
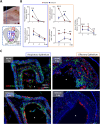
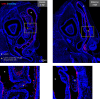
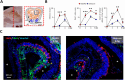
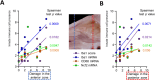
Olfactory disorders have been a hallmark of COVID-19. Since the emergence of Omicron, the prevalence of anosmia has dropped, reducing interest for Omicron pathophysiology in the nasal cavity. Using the hamster model, we show that despite a lower impact than early variants of SARS-CoV-2 (D614G), Omicron BA.1 infection leads to hyposmia as early as 2 days postinfection (dpi). While the olfactory epithelium was mostly spared from Omicron infection at 2 dpi, the respiratory epithelium was strongly infected with increased presence of inflammation markers, epithelial damage, and cellular debris in the lumen of the nasal cavity. The hyposmia caused by the early SARS-CoV-2 variant and Omicron BA.1 infection was similar at 4 and 8 dpi. At 4 dpi, Omicron successfully infected the olfactory epithelium although to a lesser extent than D614G. The BA.1 infection led to innate immune cell invasion and desquamation of the olfactory epithelium similarly as the D614G variant, with persistent inflammatory markers in the olfactory turbinates at 8 dpi despite clearance of the virus. Overall, our results indicate that Omicron successfully infects the nasal epithelium including the olfactory epithelium, but with a delay and to a lesser extent than previous SARS-CoV-2 variants. These results are consistent with Omicron pathophysiology in humans showing a decreased olfactory threshold sensitivity that may be caused, as at the early stage in hamsters, by cellular debris filling the lumen of the olfactory cleft following epithelial damage.
Keywords: olfaction; olfactory behavior; respiratory viruses.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Planas D., Saunders N., Maes P., et al., “Considerable Escape of SARS‐CoV‐2 Omicron to Antibody Neutralization,” Nature 602 (2022): 671–675. - PubMed
-
- Shuai H., Chan J. F.‐W., Hu B., et al., “Attenuated Replication and Pathogenicity of SARS‐CoV‐2 B.1.1.529 Omicron,” Nature 603 (2022): 693–699. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

