Machine Learning Reassessment of Serum Immune Factors Shows No Unique Immune Profiles Linked to Disease Outcomes in SARS-CoV-2-infected Patients at Hospital Admittance
- PMID: 40711443
- PMCID: PMC12292737
- DOI: 10.1002/eji.70001
Machine Learning Reassessment of Serum Immune Factors Shows No Unique Immune Profiles Linked to Disease Outcomes in SARS-CoV-2-infected Patients at Hospital Admittance
Abstract
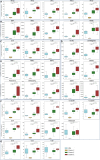
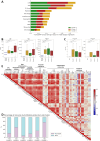
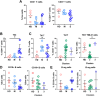
The complex pathophysiology of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) involves a hyperinflammatory state with excessive cytokine production, leading to an influenza-like syndrome that may need emergency care. The severity of SARS-CoV-2 varies widely, and collective serum immune factors, evaluated in emergency care patients, have not been shown to correlate with disease progression. We applied a machine learning approach to reassess and define serum immune profiles that could align with clinical laboratory parameters and predict disease outcomes in patients with respiratory virus infections, including those with SARS-CoV-2, seeking emergency care. Sixty-two plasma immune molecules, in a cohort of 67 symptomatic SARS-CoV-2, were analyzed for correlation with antibodies (Abs) to spike (S) and nucleocapsid (N) proteins, as well as with clinical laboratory parameters, to identify early indicators of disease prognosis at hospital admission. This approach allowed us to analyze and cluster unlabeled datasets, delineating three distinct serum immune signatures. Two showed significant and opposite modulations, correlating with poorer disease outcomes, while most patients with moderate disease displayed modest immune factor dysregulation. This highlights the complexity of immune responses in the severity of diseases caused by highly respiratory pathogenic virus like SARS-CoV-2, emphasizing the importance of evaluating overall immune imbalance rather than focusing on a few dysregulated factors.
Keywords: COVID‐19; cytokines; infection; inflammation; machine learning.
© 2025 The Author(s). European Journal of Immunology published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

