Comprehensive genomic profiling of over 10,000 advanced solid tumors
- PMID: 40711866
- PMCID: PMC12406462
- DOI: 10.18632/oncotarget.28757
Comprehensive genomic profiling of over 10,000 advanced solid tumors
Abstract
Purpose: To summarize clinically relevant genomic alterations in solid tumor samples from over 10,000 patients.
Methods: Descriptive statistics were used to summarize findings of retrospectively analyzed OncoExTra assay data from solid tumor samples.
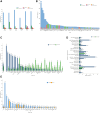
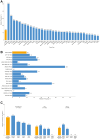
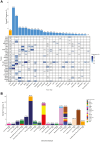
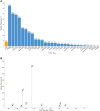
Results: The analysis cohort included 11,091 solid tumor samples from 10,768 patients. Therapeutically actionable alterations were present in 92.0% of patient samples. Biomarkers associated with on- or off-label FDA-approved therapies were detected in 29.2% and 28.0% of samples, respectively. The prevalence of hotspot alterations detected at variant allele frequency (VAF) <5% was analyzed among 7,481 samples (67.5%) harboring ≥1 of these events: 13.7% (1,022 of 7,481) had ≥1 alteration detected at VAF <5%, and 9.8% (558 of 5,690) of hotspot alterations associated with an on- or off-label FDA-approved therapy were detected at VAF <5%. Common and rare mutations in the TERT promoter were found in 8.4% (933) of samples. Whole transcriptome sequencing detected clinically relevant fusions in 7.5% of samples, with highest frequencies in prostate cancer (42.0%). The METe14 transcript was found in 14 NSCLC samples (2.7%).
Conclusions: The broad capabilities of the OncoExTra assay detected therapeutically actionable and other clinically relevant genomic events that can inform clinical decision-making for patients with advanced solid tumors.
Keywords: comprehensive genomic profiling; gene fusions; limit of detection; matched therapy; solid tumors.
Conflict of interest statement
All authors are employees of Exact Sciences.
Figures


References
-
- US Food & Drug Administration. Novel Drug Approvals at FDA. 2025. https://www.fda.gov/drugs/development-approval-process-drugs/novel-drug-....
-
- Personalized Medicine Coalition. Personalized medicine at FDA: the scope & significance of progress in 2021. 2022. https://www.personalizedmedicinecoalition.org/Userfiles/PMC-Corporate/fi....
-
- Repetto M, Fernandez N, Drilon A, Chakravarty D. Precision Oncology: 2024 in Review. Cancer Discov. 2024; 14:2332–45. 10.1158/2159-8290.CD-24-1476. - DOI - PubMed
-
- Suehnholz SP, Nissan MH, Zhang H, Kundra R, Nandakumar S, Lu C, Carrero S, Dhaneshwar A, Fernandez N, Xu BW, Arcila ME, Zehir A, Syed A, et al. Quantifying the Expanding Landscape of Clinical Actionability for Patients with Cancer. Cancer Discov. 2024; 14:49–65. 10.1158/2159-8290.CD-23-0467. - DOI - PMC - PubMed
-
- Anderson EC, DiPalazzo J, Lucas FL, Hall MJ, Antov A, Helbig P, Bourne J, Graham L, Gaitor L, Lu-Emerson C, Bradford LS, Inhorn R, Sinclair SJ, et al. Genome-matched treatments and patient outcomes in the Maine Cancer Genomics Initiative (MCGI). NPJ Precis Oncol. 2024; 8:67. 10.1038/s41698-024-00547-4. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

