Effect of Oceanic Islands on an Insect Symbiont Genome in Transition to a Host-Restricted Lifestyle
- PMID: 40711997
- PMCID: PMC12360927
- DOI: 10.1093/gbe/evaf153
Effect of Oceanic Islands on an Insect Symbiont Genome in Transition to a Host-Restricted Lifestyle
Abstract
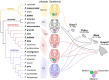
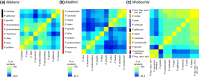
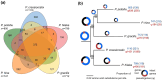
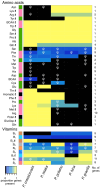
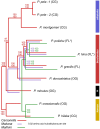
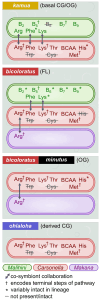
Islands offer unique opportunities to study adaptive radiations and their impacts on host genome evolution. In Hawaiian Pariaconus psyllids, all species harbor the ancient nutritional symbiont Carsonella, while only free-living and open-gall species on younger islands host a second stable cosymbiont, Makana. In contrast, a third cosymbiont, Malihini, appears to be in an early stage of host restriction and genome degradation, making it a valuable model for understanding symbiont evolution during island radiations. Here, we examine Malihini genome evolution across multiple Pariaconus lineages using 16S rRNA sequencing, metagenomics, phylogenetic reconstruction, and microscopy. We find that Malihini is codiversifying with its hosts on the oldest island Kaua'i (kamua group; open- and closed-gall makers) and on the younger islands only in free-living species (bicoloratus group). Comparison of five Malihini genomes-including three newly assembled in this study-shows ongoing genome reduction from a large-genome ancestor (>3,900 protein-coding genes), likely driven by relaxed selection, vertical transmission bottlenecks, and island dispersal over the past 5 million years. On Kaua'i, the galling psyllids appear to depend more heavily on cosymbiont (Malihini) for the biosynthesis of amino acids and B-vitamins than galling species on younger islands-especially closed-gall species, which only have Carsonella. Surprisingly, free-living psyllids on younger islands with all three symbionts show metabolic reliance similar to Kaua'i gall makers. Together, our results demonstrate that island biogeography and host plant ecology shape symbiont losses and codiversification patterns. Malihini represents an early stage of symbiont genome degradation during host restriction, in sharp contrast to its more stable coresidents, Carsonella and Makana.
Keywords: Malihini; Pariaconus; Hawaiian Islands; adaptive radiation; genome evolution; symbiosis.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Andrew S. FastQC: a quality control tool for high throughput sequence dataics—FastQC a quality control tool for high throughput sequence data. 2010. (accessed 2022 Mar 16). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Appelhans T, Detsch F, Reudenbach C, Woellauer S. mapview: interactive viewing of spatial data in R. 2018. (accessed 2024 Feb 2). https://github.com/r-spatial/mapview.
-
- Bastin S, Reyes-Betancort JA, Siverio De La Rosa F, Percy DM. Origins of the central Macaronesian psyllid lineages (Hemiptera; Psylloidea) with characterization of a new island radiation on endemic Convolvulus floridus (Convolvulaceae) in the Canary Islands. PLoS One. 2024:19:e0297062. 10.1371/journal.pone.0297062. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

