Genetically engineered chondrocyte-mimetic nanoplatform attenuates osteoarthritis by blocking IL-1β and restoring sirtuin-3
- PMID: 40712009
- PMCID: PMC12292748
- DOI: 10.1126/sciadv.adv4238
Genetically engineered chondrocyte-mimetic nanoplatform attenuates osteoarthritis by blocking IL-1β and restoring sirtuin-3
Abstract
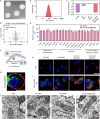
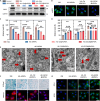
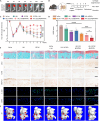
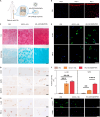
Osteoarthritis (OA) is a multifactorial disease characterized by joint inflammation and cartilage degeneration, with no disease-modifying drugs available. The vicious cycle between the inflammatory microenvironment (inflamed soil) and dysfunctional chondrocytes (degeneration-related seeds) drives the chronic progressive deterioration of OA. Here, we report a genetically engineered chondrocyte-mimetic nanoplatform (termed HKL-GECM@MPNPs) comprising a honokiol (HKL)-loaded mitochondrion-targeting nanoparticle core coated with an interleukin-1 receptor type 2 (IL-1R2)-overexpressing chondrocyte membrane. HKL-GECM@MPNPs fuse with OA chondrocytes, transferring IL-1R2 onto the plasma membrane and reprogramming the inflamed microenvironment through IL-1β blockade. Mitochondrion-targeting cores then directly deliver HKL to restore mitochondrial sirtuin-3 in OA chondrocytes, reprogramming the cells' pathological phenotype. Intra-articular injection of HKL-GECM@MPNPs in OA mice reduces inflammation, alleviates joint pain, and mitigates cartilage damage through a synergistic effect. Moreover, HKL-GECM@MPNPs effectively reverse cartilage degeneration in human OA cartilage explants. This approach highlights the potential of HKL-GECM@MPNPs to combine IL-1β blockade and mitochondrial sirtuin-3 restoration as a promising strategy for OA treatment.
Figures



References
-
- Martel-Pelletier J., Barr A. J., Cicuttini F. M., Conaghan P. G., Cooper C., Goldring M. B., Goldring S. R., Jones G., Teichtahl A. J., Pelletier J. P., Osteoarthritis. Nat. Rev. Dis. Primers. 2, 16072 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

