Hippo signaling regulates the nuclear behavior and DNA binding times of YAP and TEAD to control transcription
- PMID: 40712036
- PMCID: PMC12293108
- DOI: 10.1126/sciadv.adw4974
Hippo signaling regulates the nuclear behavior and DNA binding times of YAP and TEAD to control transcription
Abstract
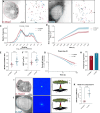
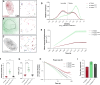
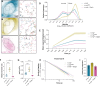
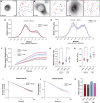
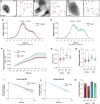
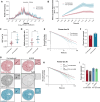
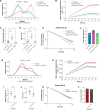
Over the past two decades, genetic and proteomic screens have identified the Hippo pathway as a complex signaling network that controls tissue growth and human cancer. Despite these advances, our understanding of how Hippo signaling regulates transcription is less clear. To address this, we used live microscopy to study the nuclear behavior of the major Hippo pathway transcription effectors, YAP and TEADs. We reveal that TEADs are a major determinant of YAP DNA binding and nuclear mobility, while YAP minorly influences TEADs. YAP and TEAD1 associate with DNA for longer periods in cells with intrinsically low Hippo pathway activity and upon acute Hippo pathway perturbation. TEAD1 binds the genome on a broad range of timescales, and this is extended substantially in nuclear condensates. Last, a cancer-associated YAP fusion protein exhibits substantially different biophysical behavior than either YAP or TEAD1. Thus, we reveal that Hippo signaling regulates transcription, in part, by influencing the DNA binding times of YAP and TEADs.
Figures
Similar articles
-
TEAD-targeting small molecules induce a cofactor switch to regulate the Hippo pathway.Proc Natl Acad Sci U S A. 2025 Jul 8;122(27):e2425984122. doi: 10.1073/pnas.2425984122. Epub 2025 Jul 3. Proc Natl Acad Sci U S A. 2025. PMID: 40608666 Free PMC article.
-
TEAD switches interacting partners along neural progenitor lineage progression to execute distinct functions.Genes Dev. 2025 Jul 1;39(13-14):849-867. doi: 10.1101/gad.352632.125. Genes Dev. 2025. PMID: 40389325
-
VGLL2 and TEAD1 fusion proteins identified in human sarcoma drive YAP/TAZ-independent tumorigenesis by engaging EP300.Elife. 2025 May 8;13:RP98386. doi: 10.7554/eLife.98386. Elife. 2025. PMID: 40338073 Free PMC article.
-
Nuclear localization of YAP in pancreatic cancer: More players besides Hippo pathway.Cancer Lett. 2025 Oct 1;629:217864. doi: 10.1016/j.canlet.2025.217864. Epub 2025 Jun 6. Cancer Lett. 2025. PMID: 40484270 Review.
-
Complex roles of Hippo-YAP/TAZ signaling in hepatocellular carcinoma.J Cancer Res Clin Oncol. 2023 Nov;149(16):15311-15322. doi: 10.1007/s00432-023-05272-2. Epub 2023 Aug 22. J Cancer Res Clin Oncol. 2023. PMID: 37608027 Free PMC article. Review.
References
-
- Gaspar P., Tapon N., Sensing the local environment: Actin architecture and Hippo signalling. Curr. Opin. Cell Biol. 31, 74–83 (2014). - PubMed
-
- Halder G., Dupont S., Piccolo S., Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 13, 591–600 (2012). - PubMed
-
- Harvey K. F., Zhang X., Thomas D. M., The Hippo pathway and human cancer. Nat. Rev. Cancer 13, 246–257 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

