DNA damage response pathway regulates Nrf2 in response to oxidative stress
- PMID: 40712037
- PMCID: PMC12293114
- DOI: 10.1126/sciadv.adu9555
DNA damage response pathway regulates Nrf2 in response to oxidative stress
Abstract
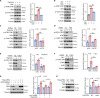
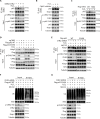
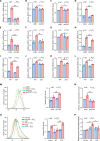
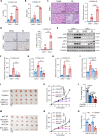
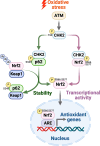
Nrf2 acts as a transcriptional master regulator to orchestrate antioxidant responses and maintain redox balance. However, the cellular pathway for translating oxidative stress signals into Nrf2-dependent antioxidant responses remain incompletely understood. Here, we show that reactive oxygen species (ROS) function as signaling molecules in modulating Nrf2's stability and transcriptional activity by activating the DNA damage response (DDR) signaling pathway. When activated, CHK2 phosphorylates the autophagy adaptor protein p62 at serine-349, promoting its interaction with Keap1 and disrupting the Keap1-Nrf2 interaction, thereby inhibiting Nrf2 ubiquitination-dependent degradation. In addition, CHK2 directly phosphorylates Nrf2 at serine-566/serine-577, enhancing its transcriptional activity and antioxidant capacity. Consistent with these effects, Chk2-/- mice show impaired expression of Nrf2 and its downstream antioxidant target genes, along with more severe renal tissue damage in an ROS-dependent model of renal ischemia/reperfusion injury. Our study reveals a direct mechanism linking the DDR signaling pathway to ROS-triggered Nrf2-dependent antioxidant responses, providing critical insight into cellular protection against oxidative stress-induced damage.
Figures


References
-
- Finkel T., Cell biology: A clean energy programme. Nature 444, 151–152 (2006). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

