Repressive Cytosine Methylation is a Marker of Viral Gene Transfer Across Divergent Eukaryotes
- PMID: 40712095
- PMCID: PMC12344493
- DOI: 10.1093/molbev/msaf176
Repressive Cytosine Methylation is a Marker of Viral Gene Transfer Across Divergent Eukaryotes
Abstract
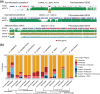
Cytosine DNA methylation patterns vary widely across eukaryotes, with its ancestral roles being understood to have included both transposable element (TE) silencing and host gene regulation. To further explore these claims, in this study, we reevaluate the evolutionary origins of DNA methyltransferases and characterize the roles of cytosine methylation on underexplored lineages, including the amoebozoan Acanthamoeba castellanii, the glaucophyte Cyanophora paradoxa, and the heterolobosean Naegleria gruberi. Our analysis of DNA methyltransferase evolution reveals a rich ancestral eukaryotic repertoire, with several eukaryotic lineages likely subsequently acquiring enzymes through lateral gene transfer (LGT). In the three species examined, DNA methylation is enriched on young TEs and silenced genes, suggesting an ancestral repressive function, without the transcription-linked gene body methylation of plants and animals. Consistent with this link with silencing, methylated genomic regions co-localize with heterochromatin marks, including H3K9me3 and H3K27me3. Notably, the closest homologs of many of the silenced, methylated genes in diverse eukaryotes belong to viruses, including giant viruses. Given the widespread occurrence of this pattern across diverse eukaryotic groups, we propose that cytosine methylation was a silencing mechanism originally acquired from bacterial donors, which was used to mitigate the expression of both transposable and viral elements, and that this function may persist in creating a permissive atmosphere for LGT in diverse eukaryotic lineages. These findings further highlight the importance of epigenetic information to annotate eukaryotic genomes, as it helps delimit potentially adaptive LGTs from silenced parasitic elements.
Keywords: DNA methylation; epigenetics; eukaryotes; heterochromatin; lateral gene transfer; viral endogenization.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Bernabeu M, Manzano-Morales S, Marcet-Houben M, Gabaldón T. Diverse ancestries reveal complex symbiotic interactions during eukaryogenesis. bioRxiv 618062. 10.1101/2024.10.14.618062, 16 October 2024, preprint: not peer reviewed. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

