A Human Immuno-Lung Organoid Model to Study Macrophage-Mediated Lung Cell Senescence Upon SARS-CoV-2 Infection
- PMID: 40712141
- PMCID: PMC12463091
- DOI: 10.1002/advs.202503932
A Human Immuno-Lung Organoid Model to Study Macrophage-Mediated Lung Cell Senescence Upon SARS-CoV-2 Infection
Abstract
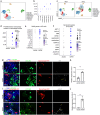
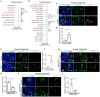
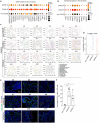
While COVID-19 affects multiple organ systems, the human respiratory system is the primary viral target and main site for disease progression. In this study, spatial transcriptional assays (NanoString CosMx) are utilized to analyze both explant and autopsy samples from non-COVID and COVID-19 lungs, identifying the activation of proinflammatory macrophages in COVID-19 explants. It is further developed immuno-lung organoids comprising hPSC-derived alveolar and airway organoids co-cultured with macrophages to investigate the impact and underlying mechanisms of macrophage-mediated lung damage following SARS-CoV-2 infection. The findings demonstrate that proinflammatory macrophages induce lung cell senescence through the THBS1-(ITGA3+ITGB1) signaling axis, a mechanism further validated using spatial transcriptomics. This study not only establishes physiologically relevant immuno-lung organoid models for modeling macrophage-mediated tissue damage, but also identifies a previous unrecognized role of the THBS1-(ITGA3+ITGB1) pathway in driving lung cell senescence during infectious disease.
Keywords: macrophage; organoid; spatial transcriptomics.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
S.C. and T.E. are the co‐founders of Oncobeat, Inc. S.C is the co‐founder of iOrganBio, Inc. The other authors have no conflict of interest.
Figures


References
-
- Kulasinghe A., Tan C. W., Miggiolaro A. F. R. S., Monkman J., SadeghiRad H., Bhuva D. D., Junior J. S. M., Paula C. B. V., Nagashima S., Baena C. P., Eur. Respir. J. 2022, 59, 2101881.
-
- Yamanaka S., Cell Stem Cell 2020, 27, 523. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
