Adaptive and metabolic convergence in rhizosphere and gut microbiomes
- PMID: 40713710
- PMCID: PMC12296625
- DOI: 10.1186/s40168-025-02179-7
Adaptive and metabolic convergence in rhizosphere and gut microbiomes
Abstract
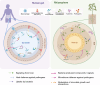
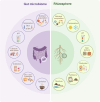
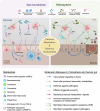
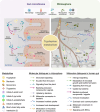
Microbial ecosystems such as the plant rhizosphere and the human gut microbiome are crucial for the health and functionality of their hosts. Despite their differences, these ecosystems share core evolutionary principles shaped by agriculture, lifestyle, and nutrient-driven selection, demonstrating resilience to environmental pressures. We introduce the concept of the human gut, particularly the colon, as an "inside-out" version of the rhizosphere, highlighting the functional and ecological parallels between the two. This review explores these analogies, focusing on metabolites and receptors involved in host-microbiome communication. By integrating insights from both ecosystems, we aim to bridge knowledge gaps and promote interdisciplinary approaches, with the potential to address global challenges in human health and agricultural sustainability. Video Abstract.
Keywords: Comparative microbiomes; Host-microbiome interactions; Human health; Microbial metabolites; Microbial resilience; Sustainable agriculture.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Nothing to declare. Consent for publication: All authors consented to publication. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Ecological mechanisms of microbial assembly in clonal plant Glechoma longituba: from soil to endosphere.Appl Environ Microbiol. 2025 Jun 18;91(6):e0033625. doi: 10.1128/aem.00336-25. Epub 2025 May 12. Appl Environ Microbiol. 2025. PMID: 40353652 Free PMC article.
-
An experimental test of the influence of microbial manipulation on sugar kelp (Saccharina latissima) supports the core influences host function hypothesis.Appl Environ Microbiol. 2025 Jun 18;91(6):e0030125. doi: 10.1128/aem.00301-25. Epub 2025 May 29. Appl Environ Microbiol. 2025. PMID: 40439420 Free PMC article.
-
Unveiling Potato Cultivars With Microbiome Interactive Traits for Sustainable Agricultural Production.Plant Cell Environ. 2025 Jul 1. doi: 10.1111/pce.70019. Online ahead of print. Plant Cell Environ. 2025. PMID: 40596781
-
Panax notoginseng-microbiota interactions: From plant cultivation to medicinal application.Phytomedicine. 2023 Oct;119:154978. doi: 10.1016/j.phymed.2023.154978. Epub 2023 Jul 17. Phytomedicine. 2023. PMID: 37549538
-
Engineering agricultural soil microbiomes and predicting plant phenotypes.Trends Microbiol. 2024 Sep;32(9):858-873. doi: 10.1016/j.tim.2024.02.003. Epub 2024 Feb 29. Trends Microbiol. 2024. PMID: 38429182 Review.
References
-
- Abdelfattah A, Wisniewski M, Schena L, Tack AJM. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environ Microbiol. 2021;23(4):2199–214. 10.1111/1462-2920.15392. - PubMed
-
- Abedini D, et al. Solanoeclepin A functions as a new signaling molecule that recruits beneficial rhizosphere microbes to promote plant growth under nitrogen deficiency. Molecular Plant. 2025. Submitted
-
- Abedini D, Jaupitre S, Bouwmeester H, Dong L. Metabolic interactions in beneficial microbe recruitment by plants. Curr Opin Biotechnol. 2021;70:241–7. 10.1016/J.COPBIO.2021.06.015. - PubMed
-
- Adolph TE, Meyer M, Schwärzler J, Mayr L, Grabherr F, Tilg H. The metabolic nature of inflammatory bowel diseases. Nat Rev Gastroenterol Hepatol. 2022;19(12):753–67. 10.1038/s41575-022-00658-y. - PubMed
-
- Agtuca BJ, Stopka SA, Tuleski TR, Do FP, Amaral SE, Liu Y, Dong Xu, Monteiro RA, Koppenaal DW, Paša-Tolić L, Anderton CR, Vertes A, Stacey G. In-situ metabolomic analysis of Setaria viridis roots colonized by beneficial endophytic bacteria. Mol Plant Microbe Interact. 2020;33(2):272–83. 10.1094/MPMI-06-19-0174-R/ASSET/IMAGES/LARGE/MPMI-06-19-0174-R_F4-1578363867287.JPEG. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

