Large MAF transcription factors reawaken evolutionarily dormant fast-glycolytic type IIb myofibers in human skeletal muscle
- PMID: 40713776
- PMCID: PMC12296675
- DOI: 10.1186/s13395-025-00391-5
Large MAF transcription factors reawaken evolutionarily dormant fast-glycolytic type IIb myofibers in human skeletal muscle
Abstract
Background: Small mammals such as mice rely on type IIb myofibers, which express the fast-contracting myosin heavy chain isoform Myh4, to achieve rapid movements. In contrast, larger mammals, including humans, have lost MYH4 expression. Thus, they favor slower-contracting myofiber types. However, the mechanisms underlying this evolutionary shift remain unclear. We recently identified the large Maf transcription factor family (Mafa, Mafb, and Maf) as key regulators of type IIb myofiber specification in mice. In this study, we investigate whether large MAFs play a conserved role in the induction of MYH4 expression and glycolytic metabolism in human and bovine skeletal muscle.
Methods: We performed adenovirus-mediated overexpression of large MAFs in iPSC-derived human myotubes and primary bovine myotubes. We subsequently quantified MYH4 expression using RT-qPCR, RNA sequencing (RNA-seq), and LC-MS/MS analysis. Glycolytic capacity was assessed using a flux analyzer and metabolic gene expression profiling. Additionally, RNA-seq analysis of human muscle biopsy samples was conducted to determine the correlations between large MAFs and the expression of MYH4 and other myosin genes, as well as their association with fast fiber composition and athletic training.
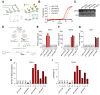
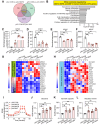
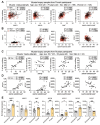
Results: Overexpression of large MAFs in human and bovine myotubes robustly induced MYH4 expression, with mRNA levels increasing by 100- to 1000-fold. LC-MS/MS analysis provided clear evidence of MYH4 protein expression in human myotubes, where it was previously undetectable. RNA-seq and flux analyzer data revealed that large MAFs significantly enhanced glycolytic capacity by upregulating the expression of key genes involved in glucose metabolism. Moreover, RNA-seq analysis of human muscle biopsy samples revealed a positive correlation between MAFA, MAF, and MYH4 expression. Furthermore, MAFA and MAF expression levels were elevated in power-trained individuals, accompanied by increased expression of MYH4 and other fast myosin genes.
Conclusions: Our findings establish large MAF transcription factors as key regulators of MYH4 expression and glycolytic metabolism in human skeletal muscle. This discovery provides novel insights into the evolutionary loss of type IIb myofibers in larger mammals and suggests potential strategies for enhancing muscle performance and mitigating fast-twitch fiber loss associated with aging and muscle degeneration.
Keywords: Aging; Human muscle; Large MAFs; MYH4; Myofiber type; Myosin heavy chain; Type IIb myofiber.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Russian part of the study was approved by the Ethics Committee of the Federal Research and Clinical Center of Physical–Chemical Medicine of the Federal Medical and Biological Agency of Russia (protocol no. 2017/04), whereas the Finnish part of the study was approved by the Hospital District of Helsinki and Uusimaa (Database of Genotypes and Phenotypes Study Accession: phs001048.v2.p1). Written informed consent was obtained from all study participants. Consent for publication: Not applicable. Competing interests: TK and ST are inventors on a patent application related to this work (Patent Application No. 2022-083553). All other authors declare they have no competing interests.
Figures


References
-
- Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol. 2013;45:2191–9. - PubMed
-
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–531. - PubMed
-
- Schiaffino S, Chemello F, Reggiani C. The diversity of skeletal muscle Fiber types. Cold Spring Harb Perspect Biol. 2024;a041477. - PubMed
-
- Albers PH, Pedersen AJT, Birk JB, Kristensen DE, Vind BF, Baba O, et al. Human muscle fiber type-specific insulin signaling: impact of obesity and type 2 diabetes. Diabetes. 2015;64:485–97. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

