Dual-targeting CSF1R signaling attenuates neurotoxic myeloid activation and preserves photoreceptors in retinitis pigmentosa
- PMID: 40713818
- PMCID: PMC12297727
- DOI: 10.1186/s12974-025-03525-0
Dual-targeting CSF1R signaling attenuates neurotoxic myeloid activation and preserves photoreceptors in retinitis pigmentosa
Abstract
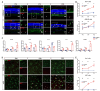
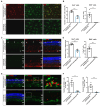
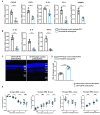
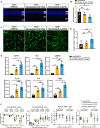
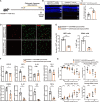
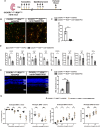
Retinitis pigmentosa (RP), a group of inherited retinal diseases characterized by progressive photoreceptor degeneration, features prominent microglial activation and monocyte-derived macrophage infiltration. While colony-stimulating factor 1 receptor (CSF1R) shows diverse roles in regulating microglial survival and behaviors in various neurodegenerative diseases, its functional significance in RP pathogenesis remains unclear. In this study, we observed upregulated CSF1R signaling specifically within disease-associated myeloid cells in the rd10 mouse model of RP. Targeted intervention via intravitreal CSF1R neutralizing antibodies and systemic PLX5622 administration achieved reduced myeloid proliferation and pro-inflammatory cytokine production and greater photoreceptor survival. Notably, CSF1R potentiation using recombinant IL-34 or CSF1 exacerbated neuroinflammation and accelerated photoreceptor degeneration. Mechanistic investigations revealed that infiltrating monocyte depletion by clodronate liposomes significantly reduced macrophage infiltration and preserved visual function. Using CX3CR1CreER/+/R26iDTR/+/rd10 mouse model, we observed that diphtheria toxin-mediated microglia ablation preserved retinal function. Overall, our findings demonstrate the prominent role of CSF1R in neurotoxic myeloid activation in the context of RP. Our results provide preclinical proof-of-concept that dual targeting of retinal and peripheral CSF1R pathways may offer a mutation-agnostic therapeutic strategy for inherited retinal degenerations.
Keywords: CSF1R; Macrophages; Microglia; Neuroinflammation; Photoreceptor degeneration; Proliferation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All experimental procedures were approved by the Animal Subjects Ethics Sub-committee (ASESC) of Hong Kong Polytechnic University and conducted in accordance with the Association for Research in Vision and Ophthalmology (ARVO) statement for the use of animals. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Microglial phagocytosis and activation underlying photoreceptor degeneration is regulated by CX3CL1-CX3CR1 signaling in a mouse model of retinitis pigmentosa.Glia. 2016 Sep;64(9):1479-91. doi: 10.1002/glia.23016. Epub 2016 Jun 17. Glia. 2016. PMID: 27314452 Free PMC article.
-
Residual microglia following short-term PLX5622 treatment in 5xFAD mice exhibit diminished NLRP3 inflammasome and mTOR signaling, and enhanced autophagy.Aging Cell. 2025 Feb;24(2):e14398. doi: 10.1111/acel.14398. Epub 2024 Nov 21. Aging Cell. 2025. PMID: 39571180 Free PMC article.
-
Understanding TAK1 deficiency in microglia: Dual mechanisms for photoreceptor protection in a mouse model of retinitis pigmentosa.Proc Natl Acad Sci U S A. 2025 May 6;122(18):e2423134122. doi: 10.1073/pnas.2423134122. Epub 2025 May 2. Proc Natl Acad Sci U S A. 2025. PMID: 40314985
-
The Relationship Between CSF1R Signaling, Monocyte-Macrophage Differentiation, and Susceptibility to Inflammatory Bowel Disease.Cell Mol Gastroenterol Hepatol. 2025;19(8):101510. doi: 10.1016/j.jcmgh.2025.101510. Epub 2025 Mar 26. Cell Mol Gastroenterol Hepatol. 2025. PMID: 40154882 Free PMC article. Review.
-
Vitamin A and fish oils for retinitis pigmentosa.Cochrane Database Syst Rev. 2013 Dec 19;2013(12):CD008428. doi: 10.1002/14651858.CD008428.pub2. Cochrane Database Syst Rev. 2013. Update in: Cochrane Database Syst Rev. 2020 Jun 18;6(6):CD008428. doi: 10.1002/14651858.CD008428.pub3. PMID: 24357340 Free PMC article. Updated.
References
-
- Dias MF, Joo K, Kemp JA, Fialho SL, da Silva Cunha A Jr., Woo SJ, et al. Molecular genetics and emerging therapies for retinitis pigmentosa: basic research and clinical perspectives. Prog Retin Eye Res. 2018;63:107–31. 10.1016/j.preteyeres.2017.10.004. - PubMed
-
- Hartong DT, Berson EL, Dryja TP, Retinitis pigmentosa. Lancet. 2006;368(9549):1795–809. 10.1016/S0140-6736(06)69740-7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

