Genes linked to schistosome resistance identified in a genome-wide association study of African snail vectors
- PMID: 40715059
- PMCID: PMC12297450
- DOI: 10.1038/s41467-025-61760-8
Genes linked to schistosome resistance identified in a genome-wide association study of African snail vectors
Abstract
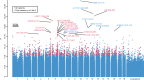
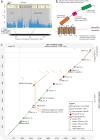
Schistosomiasis, a neglected tropical disease, is transmitted by freshwater snails. Interruption of transmission will require novel vector-focused interventions. We performed a genome-wide association study of African snails, Biomphalaria sudanica, exposed to Schistosoma mansoni in an endemic area of high transmission in Kenya. Two snail genomic regions, SudRes1 and SudRes2, were significantly associated with snail resistance to schistosomes. SudRes1 includes receptor-like protein tyrosine phosphatases while SudRes2 includes a class of leucine-rich repeat-containing G-protein coupled receptors, both comprising diverse extracellular binding domains suggestive of host-pathogen interaction. Resistant and susceptible haplotypes show numerous coding differences including presence/absence of entire genes. No loci previously tied to schistosome resistance in a neotropical snail species showed any association with compatibility suggesting that loci involved in the resistance of African vectors are distinct. Snail ancestry was also strongly correlated with parasite compatibility. These results will inform future efforts to predict and manipulate immunity of a major schistosome vector.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests
Figures


Update of
-
Immune targets for schistosomiasis control identified by a genome-wide association study of East African snail vectors.bioRxiv [Preprint]. 2024 Sep 2:2024.08.30.610565. doi: 10.1101/2024.08.30.610565. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 27;16(1):6918. doi: 10.1038/s41467-025-61760-8. PMID: 39282449 Free PMC article. Updated. Preprint.
-
Immune targets for schistosomiasis control identified by a genome-wide association study of African snail vectors.Res Sq [Preprint]. 2025 Jan 3:rs.3.rs-5656395. doi: 10.21203/rs.3.rs-5656395/v1. Res Sq. 2025. Update in: Nat Commun. 2025 Jul 27;16(1):6918. doi: 10.1038/s41467-025-61760-8. PMID: 39801518 Free PMC article. Updated. Preprint.
References
-
- World Health Organization (WHO) Global report on neglected tropical diseases. WHO 2024: Geneva, Switzerland (2024).
-
- Hay, S. I. et al. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet390, 1260–1344 (2017). - PMC - PubMed
-
- World Health Organization (WHO), WHO guideline on control and elimination of human schistosomiasis. (WHO 2022, Geneva, Switzerland, accessed 26 June 2025); https://iris.who.int/handle/10665/351856. - PubMed
-
- World Health Organization (WHO), Ending the neglect to attain the Sustainable Development Goals – A road map for neglected tropical diseases 2021–2030. (WHO 2020, Geneva, Switzerland, accessed 26 June 2025); https://iris.who.int/handle/10665/338565.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

