Large-scale RNA-Seq mining reveals ciclopirox olamine induces TDP-43 cryptic exons
- PMID: 40715064
- PMCID: PMC12297563
- DOI: 10.1038/s41467-025-62004-5
Large-scale RNA-Seq mining reveals ciclopirox olamine induces TDP-43 cryptic exons
Abstract
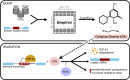
Nuclear clearance and cytoplasmic aggregation of TDP-43, initially identified in ALS-FTD, are hallmark pathological features observed across a spectrum of neurodegenerative diseases. We previously found that TDP-43 loss-of-function leads to transcriptome-wide inclusion of deleterious cryptic exons, a signature detected in presymptomatic biofluids and postmortem ALS-FTD brain tissue, but the upstream mechanisms that lead to TDP-43 dysregulation remain unclear. Here, we developed a web-based resource (SnapMine) to determine the levels of TDP-43 cryptic exon inclusion across hundreds of thousands of publicly available RNA sequencing datasets. We established cryptic exon inclusion levels across a variety of human cells and tissues to provide ground truth references for future studies on TDP-43 dysregulation. We then explored studies that were entirely unrelated to TDP-43 or neurodegeneration and found that ciclopirox olamine (CPX), an FDA-approved antifungal, can trigger the inclusion of TDP-43-associated cryptic exons in a variety of mouse and human primary cells. CPX induction of cryptic exons arises from heavy metal toxicity and oxidative stress, suggesting that similar vulnerabilities could play a role in neurodegeneration. Our work demonstrates how diverse datasets can be linked through common biological features and underscores how public archives of sequencing data remain a vastly underutilized resource with tremendous potential for uncovering novel insights into complex biological mechanisms and diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
Large-scale RNA-seq mining reveals ciclopirox triggers TDP-43 cryptic exons.bioRxiv [Preprint]. 2024 Mar 30:2024.03.27.587011. doi: 10.1101/2024.03.27.587011. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 25;16(1):6878. doi: 10.1038/s41467-025-62004-5. PMID: 38585725 Free PMC article. Updated. Preprint.
References
-
- Wahl, M. C., Will, C. L. & Lührmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell136, 701–718 (2009). - PubMed
-
- Matlin, A. J., Clark, F. & Smith, C. W. J. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol.6, 386–398 (2005). - PubMed
-
- Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet.40, 1413–1415 (2008). - PubMed
MeSH terms
Substances
Grants and funding
- UH3NS115608/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R33NS115161/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OAC1920103/National Science Foundation (NSF)
- 1U01FD008129/U.S. Department of Health & Human Services | U.S. Food and Drug Administration (U.S. Food & Drug Administration)
- R01 NS127186/NS/NINDS NIH HHS/United States
- R33 NS115161/NS/NINDS NIH HHS/United States
- R01NS127186/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01NS095969/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 NS095969/NS/NINDS NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- U01 FD008129/FD/FDA HHS/United States
- UH3 NS115608/NS/NINDS NIH HHS/United States
- DGE2139757/National Science Foundation (NSF)
LinkOut - more resources
Full Text Sources
Miscellaneous

