Melbournevirus encodes a shorter H2B-H2A doublet histone variant that forms structurally distinct nucleosome structures
- PMID: 40715086
- PMCID: PMC12297534
- DOI: 10.1038/s41467-025-62031-2
Melbournevirus encodes a shorter H2B-H2A doublet histone variant that forms structurally distinct nucleosome structures
Abstract
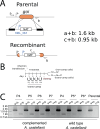
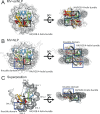
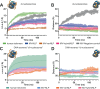
Unique among viruses, some giant viruses utilize histones to organize their genomes into nucleosomes. Melbournevirus encodes a distinct H2B-H2A histone doublet variant in addition to the canonical H4-H3 and H2B-H2A doublets. This viral histone variant has a truncated H2B portion and its amino acid sequence deviates from that of the main viral H2B-H2A throughout the entire coding region. It is less abundant than the main H2B-H2A doublet, is likely essential for melbournevirus fitness, and is conserved in all Marseilleviridae. The cryo-EM structure of a nucleosome-like particle reconstituted with this H2B-H2A variant and viral H4-H3 reveals that only 90 base pairs of DNA are stably bound, significantly less than in eukaryotic nucleosomes and viral nucleosomes made with the main fused viral histone doublets. The reduced ability to bind DNA can be attributed to structural differences between variant and main H2B-H2A. Variant melbournevirus nucleosomes are less stable, possibly aiding rapid genome unpacking to initiate gene expression. Our results highlight the remarkable propensity of giant viruses to appropriate the utility of histones for their specialized purposes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature389, 251–260 (1997). - PubMed
-
- Stevens, K. M. & Warnecke, T. Histone variants in archaea – An undiscovered country. Semin. Cell Dev. Biol.135, 50–58 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

