Biomarkers of pediatric Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis through single-cell transcriptomics
- PMID: 40715093
- PMCID: PMC12297430
- DOI: 10.1038/s41467-025-62090-5
Biomarkers of pediatric Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis through single-cell transcriptomics
Abstract
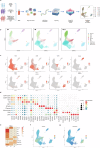
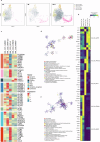
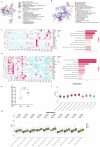
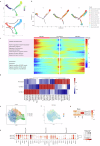
Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis (EBV-HLH) is a fatal hyperinflammatory disorder distinct from self-limiting EBV-induced infectious mononucleosis (IM). However, the immunological mechanisms underlying the divergence between benign EBV infection and fulminant HLH-particularly in the absence of inherited immunodeficiency-remains unclear, and systematic comparisons of immune landscapes across EBV-associated disease spectra are lacking. In this study, by enrolling children with IM and healthy volunteers as controls, we utilize single-cell RNA sequencing to identify unique immunological characteristics of EBV-HLH. Our analysis indicates that patients with EBV-HLH exhibite widespread activation of NF-κB signaling pathway. Furthermore, excessive cytokine secretion by T and NK cells is observed, along with a shift in monocyte differentiation towards an inflammatory phenotype, and the aggregation of IDO1+ monocytes. Metabolic pathway analysis reveals that L-kynurenine, a downstream metabolite of IDO1, is specifically elevated in EBV-HLH and mediates the production of multiple pro-inflammatory cytokines. Collectively, our study maps the immune landscape in pediatric EBV-HLH at single-cell resolution, uncovering potential role of IDO1+ monocytes and L-kynurenine as biomarkers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Epstein, M. A., Achong, B. G. & Barr, Y. M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet1, 702–703 (1964). - PubMed
-
- Taylor, G. S., Long, H. M., Brooks, J. M., Rickinson, A. B. & Hislop, A. D. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol.33, 787–821 (2015). - PubMed
-
- El-Mallawany, N. K., Curry, C. V. & Allen, C. E. Haemophagocytic lymphohistiocytosis and Epstein-Barr virus: a complex relationship with diverse origins, expression and outcomes. Br. J. Haematol.196, 31–44 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

