The mechanotransducer Piezo1 coordinates metabolism and inflammation to promote skin growth
- PMID: 40715139
- PMCID: PMC12297709
- DOI: 10.1038/s41467-025-62270-3
The mechanotransducer Piezo1 coordinates metabolism and inflammation to promote skin growth
Abstract
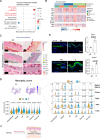
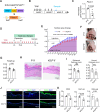
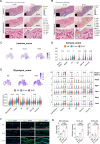
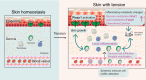
The skin has a remarkable ability to grow under constant stretch. Using a controlled tissue expansion system in mice, we identified an enhanced inflammatory-metabolic network in stretched skin via single-cell RNA sequencing, flow cytometry and spatial transcriptomics. Stretched epidermal cells exhibit heightened cellular crosstalk of CXCL, CCL, TNF, and TGF-β signaling. Additionally, skin expansion increases macrophage and monocyte infiltration in the skin while altering systemic immune cell profiles. Glycolysis-related genes, including Glut1 and Aldoa were significantly elevated. We hypothesize that Piezo1, a non-selective calcium-permeable cation channel, senses tension in stretched skin, driving these responses. The epidermal-Piezo1 loss-of-function animals show reduced skin growth, tissue weight, tissue thickness, macrophage infiltration, and glycolysis activity. Conversely, animals with a pharmacological Piezo1 gain of function exhibit an increase in these factors. Our findings highlight the coordinating role of Piezo1 for metabolic changes and immune cell infiltration in tension-induced skin growth.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: L.A.G. has received grant support paid to his institution, Johns Hopkins University from Sun Pharma Advanced Research Company (SPARC). This grant is to investigate intellectual property where Johns Hopkins University is the owner, L.A.G. is one of several inventors, is under a licensing agreement with SPARC, and has resulted in royalty payments to inventors. This grant and royalty payments are not related to the research presented in this manuscript. The remaining authors declare no competing interests.
Figures



References
-
- Radovan, C. Tissue expansion in soft-tissue reconstruction. Plast. Reconstr. Surg.74, 482–492 (1984). - PubMed
-
- De Filippo, R. E. & Atala, A. Stretch and growth: the molecular and physiologic influences of tissue expansion. Plast. Reconstr. Surg.109, 2450–2462 (2002). - PubMed
-
- Takei, T., Mills, I., Arai, K. & Sumpio, B. E. Molecular basis for tissue expansion: clinical implications for the surgeon. Plast. Reconstr. Surg.102, 247–258 (1998). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

