Regulation of RPE65 expression in human retinal pigment epithelium cells
- PMID: 40715346
- PMCID: PMC12297382
- DOI: 10.1038/s41598-025-12926-3
Regulation of RPE65 expression in human retinal pigment epithelium cells
Abstract
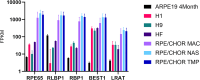
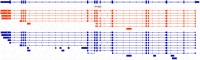
The visual cycle is an important pathway in the retinal pigment epithelium (RPE) which regenerates 11-cis retinal chromophore for the retinal photoreceptors. The central enzyme in the visual cycle is RPE65 retinol isomerase. Expression of RPE65 mRNA and protein levels are significantly lower in RPE cell culture models when compared to native RPE. This limits the use of these models to study the visual cycle. To determine the main drivers of RPE65 regulation we compared the transcriptional profiles of native and cell culture models of RPE with various levels of RPE65 expression. We also compared the levels of RPE65 expression between ARPE-19 cells grown in media supplemented with 1 mM pyruvate (PYR) or 10 mM nicotinamide (NAM). In addition, we performed experiments directed at transcriptional and translational regulation of RPE65. We show that RPE65 mRNA and protein expression is significantly higher in NAM media grown cells than PYR cells. Transfection of cells with a variety of different vectors containing RPE65 ORFs with different promoters, codon optimization, IRES, 3' UTRs, suggest that translational effects are less important than transcriptional status. Importantly, we found that feeding with rod outer segments (ROS) decreases RPE65 expression in NAM grown cells, suggesting that certain primary functions of the RPE (here, visual cycle and phagocytosis) are not positively linked. Analysis of differentially regulated microRNAs (miRs) provides a basis for this downregulation. It appears that the regulation of RPE65 expression in ARPE-19 cells, in particular, is multifactorial, involving primarily metabolic and transcriptional status of the cells, with translation of RPE65 mRNA playing a smaller role.
Keywords: MicroRNAs; Nicotinamide; Pyruvate; RPE65; Retina; Retinal pigment epithelium; Ribosome; Transcription; Translation.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
Peripheral iridotomy for pigmentary glaucoma.Cochrane Database Syst Rev. 2016 Feb 12;2(2):CD005655. doi: 10.1002/14651858.CD005655.pub2. Cochrane Database Syst Rev. 2016. PMID: 26871761 Free PMC article.
-
A History of the Classical Visual Cycle.Prog Mol Biol Transl Sci. 2015;134:433-48. doi: 10.1016/bs.pmbts.2015.06.009. Epub 2015 Jul 3. Prog Mol Biol Transl Sci. 2015. PMID: 26310169 Review.
-
Epidemiology of Mutations in the 65-kDa Retinal Pigment Epithelium (RPE65) Gene-Mediated Inherited Retinal Dystrophies: A Systematic Literature Review.Adv Ther. 2022 Mar;39(3):1179-1198. doi: 10.1007/s12325-021-02036-7. Epub 2022 Jan 30. Adv Ther. 2022. PMID: 35098484 Free PMC article.
-
Interventions for central serous chorioretinopathy: a network meta-analysis.Cochrane Database Syst Rev. 2025 Jun 16;6(6):CD011841. doi: 10.1002/14651858.CD011841.pub3. Cochrane Database Syst Rev. 2025. PMID: 40522203
-
Prime Editing Strategy to Install the RPE65 c.1430A>G Dominant Mutation.Adv Exp Med Biol. 2025;1468:101-106. doi: 10.1007/978-3-031-76550-6_17. Adv Exp Med Biol. 2025. PMID: 39930180
References
-
- Hamel, C. P. et al. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem.268, 15751–15757 (1993). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

