The role of glucocorticoids in hepatocellular carcinoma through integrated bioinformatics analysis and experimental validation
- PMID: 40715580
- PMCID: PMC12297655
- DOI: 10.1038/s41598-025-12839-1
The role of glucocorticoids in hepatocellular carcinoma through integrated bioinformatics analysis and experimental validation
Abstract
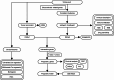
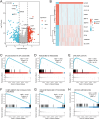
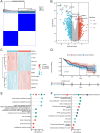
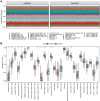
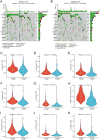
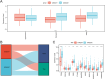
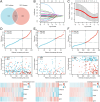
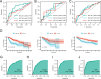
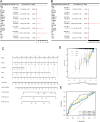
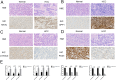
The most widespread primary liver cancer around the world is hepatocellular carcinoma (HCC), and its rising incidence and mortality rates are major challenges for public health. This study investigates the role of glucocorticoids in HCC, focusing on their associated phenotypic genes and their impact on patient prognosis. Utilizing comprehensive bioinformatics approaches, a total of 751 differentially expressed genes were identified, with 470 showing increased expression and 281 showing decreased expression in tumor samples. Gene set enrichment analysis (GSEA) indicated that tumor samples showed significant enrichment in spliceosome, ribosome, and DNA replication pathways, while control samples were enriched in complement and coagulation cascades and drug metabolism pathways. Furthermore, based on glucocorticoid-related genes, consensus clustering categorized HCC samples into two subtypes, with subtype 2 exhibiting poorer prognosis. Immune infiltration analysis indicated significant differences in various immune cell types between the two subtypes, suggesting potential immune evasion mechanisms. Drug sensitivity analysis from the Genomics of Drug Sensitivity in Cancer (GDSC) database revealed that subtype 2 patients may be more responsive to certain drugs, such as Bortezomib and Dactinomycin. Furthermore, based on the established prognostic model, a total of four genes (KIF2C, CYP2C9, PON1, SPP1) were identified. These genes are both glucocorticoid-related receptors and closely associated with the development of hepatocellular carcinoma, and they have reliable diagnostic and prognostic value. DGIdb drug prediction shows that a variety of drugs and compounds can target these four genes. Finally, immunohistochemistry revealed that in contrast to normal liver tissues, KIF2C and SPP1 were highly expressed in tumor tissues, while CYP2C9 and PON1 were expressed at lower levels in tumor tissues. This study highlights the importance of glucocorticoid-related genes in the development and prognosis of HCC, providing insights for future experimental validation and clinical applications.
Keywords: Bioinformatics; Glucocorticoid-related genes; Glucocorticoids; Hepatocellular carcinoma; Prognostic genes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

