Deep graph convolutional network-based multi-omics integration for cancer driver gene identification
- PMID: 40716043
- PMCID: PMC12296362
- DOI: 10.1093/bib/bbaf364
Deep graph convolutional network-based multi-omics integration for cancer driver gene identification
Abstract
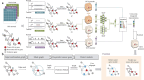
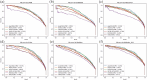
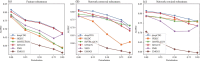
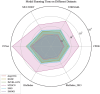
Cancer driver genes play a pivotal role in understanding cancer development, progression, and therapeutic discovery. The plenty of accumulation of multi-omics data and biological networks provides a data foundation for graph neural network (GNN) frameworks. However, most existing methods directly concatenate multi-omics data as features, which may lead to limited performance. To address this limitation, we propose deepCDG, a deep graph convolutional network (GCN)-based multi-omics integration model for cancer driver gene identification. The model first employs shared-parameter GCN encoders to extract representations from three omics perspectives, followed by feature integration through an attention layer, and finally utilizes a residual-connected GCN predictor for cancer driver gene identification. Additionally, deepCDG employs GNNExplainer for cancer driver gene module identification. Experimental results demonstrate the effective predictive performance, model robustness, and computational efficiency of deepCDG. Additionally, biological interpretability analysis further validates the reliability of the identification of cancer driver genes of our framework, and the identified gene modules provide profound insights into complex inter-gene relationships and interactions. We believe our method offers enhanced applicability for cancer driver gene identification and could be extended to other biological research fields in future studies.
Keywords: cancer driver genes; gene modules; graph convolutional networks; multi-omics data.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


Similar articles
-
DriverOmicsNet: an integrated graph convolutional network for multi-omics exploration of cancer driver genes.Brief Bioinform. 2025 Jul 2;26(4):bbaf412. doi: 10.1093/bib/bbaf412. Brief Bioinform. 2025. PMID: 40814229 Free PMC article.
-
MO-GCAN: multi-omics integration based on graph convolutional and attention networks.Bioinformatics. 2025 Aug 2;41(8):btaf405. doi: 10.1093/bioinformatics/btaf405. Bioinformatics. 2025. PMID: 40692180 Free PMC article.
-
Interpretable graph Kolmogorov-Arnold networks for multi-cancer classification and biomarker identification using multi-omics data.Sci Rep. 2025 Jul 29;15(1):27607. doi: 10.1038/s41598-025-13337-0. Sci Rep. 2025. PMID: 40730661 Free PMC article.
-
Graph neural networks for single-cell omics data: a review of approaches and applications.Brief Bioinform. 2025 Mar 4;26(2):bbaf109. doi: 10.1093/bib/bbaf109. Brief Bioinform. 2025. PMID: 40091193 Free PMC article.
-
Deep learning-driven multi-omics analysis: enhancing cancer diagnostics and therapeutics.Brief Bioinform. 2025 Jul 2;26(4):bbaf440. doi: 10.1093/bib/bbaf440. Brief Bioinform. 2025. PMID: 40874818 Free PMC article. Review.
References
-
- Li X, Hao J, Zhao Z. et al. PathActMarker: an R package for inferring pathway activity of complex diseases. Front Comp Sci 2025;19:193908.
MeSH terms
Grants and funding
- 62202383/National Natural Science Foundation of China
- 62433016/National Natural Science Foundation of China
- 2024A1515012602/Guangdong Basic and Applied Basic Research Foundation
- 2022YFD1801200/National Key Research and Development Program of China
- SKLADCPKFKT202407/State Key Laboratory for Animal Disease Control and Prevention Foundation
LinkOut - more resources
Full Text Sources
Medical

