Multiplexed Chromatin Analysis Using Optical Spectroscopic Statistical Nanosensing
- PMID: 40718523
- PMCID: PMC12288678
- DOI: 10.1021/acsphotonics.5c00311
Multiplexed Chromatin Analysis Using Optical Spectroscopic Statistical Nanosensing
Abstract
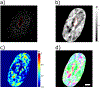
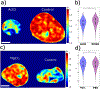
In single cells, chromatin packs into organized structures to perform biological functions, such as RNA transcription regulation. Characterizing such structural behaviors, including packing density and mass scaling, is critical in epigenetics research. Partial wave spectroscopic (PWS) microscopy is a label-free, live-cell, high-throughput imaging modality that utilizes optical spectroscopic statistical nanosensing. Rather than resolving the exact chromatin packing structure, PWS extracts statistical packing information from spectroscopic interference signals. In this study, we evaluate its ability to characterize multiplexed chromatin packing density and mass scaling, as well as its spatial confidence interval, using finite difference time domain (FDTD) electromagnetic simulations. We validated the simulation-based analysis algorithm by comparing experimental PWS images against coregistered super-resolution acquisitions, confirming its accuracy in capturing chromatin packing metrics. We then applied this modality to live cells treated with different epigenetic agents, mapping spatial changes in chromatin packing in a high-throughput workflow.
Keywords: chromatin imaging; finite difference time domain simulation; spectroscopic nanosensing.
Figures






Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
A New Measure of Quantified Social Health Is Associated With Levels of Discomfort, Capability, and Mental and General Health Among Patients Seeking Musculoskeletal Specialty Care.Clin Orthop Relat Res. 2025 Apr 1;483(4):647-663. doi: 10.1097/CORR.0000000000003394. Epub 2025 Feb 5. Clin Orthop Relat Res. 2025. PMID: 39915110
-
Artificial intelligence for diagnosing exudative age-related macular degeneration.Cochrane Database Syst Rev. 2024 Oct 17;10(10):CD015522. doi: 10.1002/14651858.CD015522.pub2. Cochrane Database Syst Rev. 2024. PMID: 39417312
-
Novel application of metabolic imaging of early embryos using a light-sheet on-a-chip device: a proof-of-concept study.Hum Reprod. 2025 Jan 1;40(1):41-55. doi: 10.1093/humrep/deae249. Hum Reprod. 2025. PMID: 39521726 Free PMC article.
-
Tobacco packaging design for reducing tobacco use.Cochrane Database Syst Rev. 2017 Apr 27;4(4):CD011244. doi: 10.1002/14651858.CD011244.pub2. Cochrane Database Syst Rev. 2017. PMID: 28447363 Free PMC article.
References
-
- Almassalha LM; Bauer GM; Chandler JE; Gladstein S; Cherkezyan L; Stypula-Cyrus Y; Weinberg S; Zhang D; Thusgaard Ruhoff P; Roy HK; Subramanian H; Chandel NS; Szleifer I; Backman V Label-free imaging of the native, living cellular nanoarchitecture using partial-wave spectroscopic microscopy. Proc. Natl. Acad. Sci. U. S. A 2016, 113, E6372–E6381. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
