Strategies for Competitive Activity-Based Protein Profiling in Small Molecule Inhibitor Discovery and Characterization
- PMID: 40718598
- PMCID: PMC12291166
- DOI: 10.1002/ijch.202200113
Strategies for Competitive Activity-Based Protein Profiling in Small Molecule Inhibitor Discovery and Characterization
Abstract
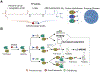
Since its introduction by Cravatt and colleagues in 1999, activity-based protein profiling (ABPP) has become widely utilized throughout academia, government, and industry laboratories to study enzymes spanning numerous gene families in a multitude of biological systems. As a variation of ABPP, competitive ABPP provides a powerful approach to characterize the binding behavior of small molecule probes and clinical drugs throughout the functional proteome. The power and flexibility of competitive ABPP are exemplified by a wide range of creative adaptions which increase assay throughput, enable diverse detection schemes, and support the implementation of this approach within a hybrid target-based screening platform. We review major developments in competitive ABPP through a compare-contrast format to provide a useful introduction to this enabling technology for scientists in chemical biology and drug discovery.
Figures
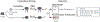



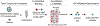

Similar articles
-
Primary Amine-Based Photoclick Chemistry: From Concept to Diverse Applications in Chemical Biology and Medicinal Chemistry.Acc Chem Res. 2025 Jul 1;58(13):1963-1981. doi: 10.1021/acs.accounts.5c00158. Epub 2025 Jun 18. Acc Chem Res. 2025. PMID: 40532071 Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Variation within and between digital pathology and light microscopy for the diagnosis of histopathology slides: blinded crossover comparison study.Health Technol Assess. 2025 Jul;29(30):1-75. doi: 10.3310/SPLK4325. Health Technol Assess. 2025. PMID: 40654002 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
