Integrating multi-dimensional data to reveal the mechanisms and molecular targets of baikening granules for treatment of pediatric influenza
- PMID: 40718794
- PMCID: PMC12289495
- DOI: 10.3389/fmolb.2025.1637980
Integrating multi-dimensional data to reveal the mechanisms and molecular targets of baikening granules for treatment of pediatric influenza
Abstract
Background: Children are the main group affected by the influenza virus, posing challenges to their health. The high risk of viral variability, drug resistance, and drug development leads to a scarcity of therapeutic drugs. Baikening (BKN) granules are a marketed traditional Chinese medicine used to treat children's lung heat, asthma, whooping cough, etc. Therefore, exploring the potential mechanisms of BKN in treating pediatric influenza is of great significance for discovering new drugs.
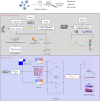
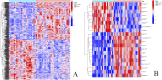
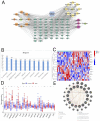
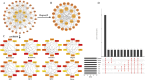
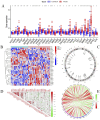
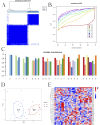
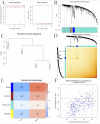
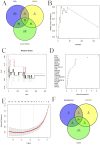
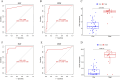
Methods: Through the database, we obtained differentially expressed genes (DEGs) between pediatric influenza and healthy samples, identified the components of BKN, and collected the targets. Target networks were built with the purpose of screening both targets and key components. Pathway and function enrichment were conducted on the relevant targets of BKN for treating pediatric influenza. BKN-related hub genes for influenza were discovered through DEGs, weighted gene co-expression network analysis (WGCNA), BKN-cluster WGCNA, and machine learning model. The accuracy of prediction efficiency and the value of BKN-related hub gene were validated through analysis of external datasets and receiver operating characteristics. Ultimately, simulations using molecular docking and molecular dynamics were used to forecast how active components will bind to hub genes.
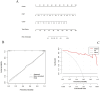
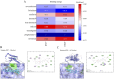
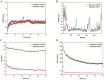
Result: A total of 20 candidate active compounds, 58 potential targets, and 3,819 DEGs were identified. The target network screened the top 10 key components and 6 core targets (PPARG, MMP2, GSK3B, PARP1, CCNA2, and IGF1). Potential target enrichment analysis indicated that BKN may be involved in AMPK signaling pathway, PI3K Akt signaling pathway, etc., to combat pediatric influenza. Subsequently, two hub genes (OTOF, IFI27) were obtained through WGCNA, BKN-cluster WGCNA, and machine learning models as potential biomarkers for BKN-related pediatric influenza. Two hub genes were found to have primary diagnostic value based on ROC curve analysis. Molecular docking confirmed the binding between BKN and hub gene. Molecular dynamics further revealed the stable binding between Peimisine and hub genes.
Conclusion: BKN may alleviate pediatric influenza via key components targeting core targets (PPARG, MMP2, GSK3B, PARP1, CCNA2, and IGF1) and hub genes (OTOF, IFI27), with the involvement of feature genes-related pathways. These results have potential consequences for future research and clinical practice.
Keywords: baikening granules; bioinformatics; machine learning; molecular docking; molecular dynamics simulation; network pharmacology; pediatric influenza.
Copyright © 2025 Gong, Che, Hu, Song, Chen, Zhang, Liang, Li, Zhao, Yan, Zhang, Liu, Guo and Shi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Elucidating the Mechanism of Xiaoqinglong Decoction in Chronic Urticaria Treatment: An Integrated Approach of Network Pharmacology, Bioinformatics Analysis, Molecular Docking, and Molecular Dynamics Simulations.Curr Comput Aided Drug Des. 2025 Jul 16. doi: 10.2174/0115734099391401250701045509. Online ahead of print. Curr Comput Aided Drug Des. 2025. PMID: 40676786
-
Integrated bioinformatics and network pharmacology to identify and validate macrophage polarization related hub genes in the treatment of osteoarthritis with Astragalus membranaceus.J Orthop Surg Res. 2025 May 30;20(1):543. doi: 10.1186/s13018-025-05799-9. J Orthop Surg Res. 2025. PMID: 40442788 Free PMC article.
-
Understanding mechanisms of Polygonatum sibiricum-derived exosome-like nanoparticles against breast cancer through an integrated metabolomics and network pharmacology analysis.Front Chem. 2025 Jun 6;13:1559758. doi: 10.3389/fchem.2025.1559758. eCollection 2025. Front Chem. 2025. PMID: 40547857 Free PMC article.
-
Medicine for chronic atrophic gastritis: a systematic review, meta- and network pharmacology analysis.Ann Med. 2023;55(2):2299352. doi: 10.1080/07853890.2023.2299352. Epub 2024 Jan 3. Ann Med. 2023. PMID: 38170849 Free PMC article.
-
Clinical symptoms, signs and tests for identification of impending and current water-loss dehydration in older people.Cochrane Database Syst Rev. 2015 Apr 30;2015(4):CD009647. doi: 10.1002/14651858.CD009647.pub2. Cochrane Database Syst Rev. 2015. PMID: 25924806 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

