Prevalence of antibiotic resistance genes its association with microbiota in raw milk of northwest Xinjiang
- PMID: 40718809
- PMCID: PMC12289685
- DOI: 10.3389/fmicb.2025.1595051
Prevalence of antibiotic resistance genes its association with microbiota in raw milk of northwest Xinjiang
Abstract
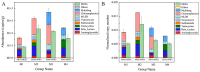
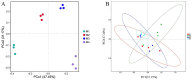
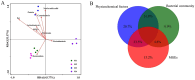
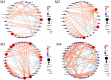
The issue of antibiotic resistance caused by antibiotic resistance genes (ARGs) has become a significant concern in environmental research in recent years, while raw milk is an important link in the food chain and has become one of the carriers and reservoirs of ARGs, which has not been taken seriously. This research employed high-throughput quantitative PCR and Illumina sequencing techniques targeting the 16S rRNA gene. These methods were used to examine the bacterial community composition and genes associated with antibiotic resistance in raw milk samples collected from the northwestern area of Xinjiang. An aggregate of 31 distinct resistance alleles were identified, with their abundance reaching as high as 3.70 × 105 copies per gram in the analyzed raw milk samples. Microorganisms harboring ARGs that confer resistance to beta-lactams, tetracyclines, aminoglycosides, and chloramphenicol derivatives were prevalent in raw milk. Procrustes analysis revealed a certain degree of correlation between the microbial community and the antibiotic resistance gene (ARG) profiles. Furthermore, network analysis demonstrated that Actinobacteria and Firmicutes were the predominant phyla exhibiting co-occurrence relationships with specific ARGs. Combining the findings from Variance Partitioning Analysis (VPA), the distribution of ARGs was mainly driven by three factors: the combined effect of physicochemical properties and mobile genetic elements (MGEs) (33.5%), the interplay between physicochemical parameters and microbial communities (31.8%), and the independent contribution of physicochemical factors (20.7%). The study demonstrates that the overall abundance of ARGs correlates with physicochemical parameters, bacterial community composition, and the presence of MGEs. Furthermore, understanding these associations facilitates the evaluation of antibiotic resistance risks, thereby contributing to enhanced farm management practices and the assurance of food safety.
Keywords: High throughput qPCR; antibiotic resistance genes; antibiotic-resistant bacteria; host; raw milk.
Copyright © 2025 Zhao, Niu, Zhao, Huang and Qin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Distributions of pathogenic bacteria, antibiotic resistance genes, and virulence factors in pig farms in China.Ecotoxicol Environ Saf. 2025 Sep 1;302:118607. doi: 10.1016/j.ecoenv.2025.118607. Epub 2025 Jul 4. Ecotoxicol Environ Saf. 2025. PMID: 40609272
-
The transfer of antibiotic resistance genes between evolutionarily distant bacteria.mSphere. 2025 Jun 25;10(6):e0011425. doi: 10.1128/msphere.00114-25. Epub 2025 Jun 3. mSphere. 2025. PMID: 40459279 Free PMC article.
-
[Occurrence Characteristics and Consumption Risk of Antibiotic Resistance Genes in Organic Vegetables].Huan Jing Ke Xue. 2025 Jul 8;46(7):4723-4732. doi: 10.13227/j.hjkx.202406155. Huan Jing Ke Xue. 2025. PMID: 40677084 Chinese.
-
The neonatal intestinal resistome and factors that influence it-a systematic review.Clin Microbiol Infect. 2022 Dec;28(12):1539-1546. doi: 10.1016/j.cmi.2022.07.014. Epub 2022 Jul 19. Clin Microbiol Infect. 2022. PMID: 35868586
-
Prophylactic antibiotics for adults with chronic obstructive pulmonary disease: a network meta-analysis.Cochrane Database Syst Rev. 2021 Jan 15;1(1):CD013198. doi: 10.1002/14651858.CD013198.pub2. Cochrane Database Syst Rev. 2021. PMID: 33448349 Free PMC article.
Cited by
-
Staphylococci in Livestock: Molecular Epidemiology, Antimicrobial Resistance, and Translational Strategies for One Health Protection.Vet Sci. 2025 Aug 13;12(8):757. doi: 10.3390/vetsci12080757. Vet Sci. 2025. PMID: 40872707 Free PMC article. Review.
References
-
- Chen B., Grandison A. S., Lewis M. J. (2017). Best use for milk - A review. II - Effect of physiological, husbandry and seasonal factors on the physicochemical properties of bovine milk. Int. J. Dairy Technol. 70, 1–15. 10.1111/1471-0307.12355 - DOI
LinkOut - more resources
Full Text Sources

