Capsaicin Alleviates Autophagy-Lysosomal Dysfunction via PPARA-Mediated V-ATPase Subunit ATP6V0E1 Signaling in 3xTg-AD Mice
- PMID: 40719096
- PMCID: PMC12533174
- DOI: 10.1002/advs.202502707
Capsaicin Alleviates Autophagy-Lysosomal Dysfunction via PPARA-Mediated V-ATPase Subunit ATP6V0E1 Signaling in 3xTg-AD Mice
Abstract
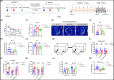
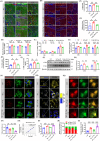
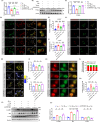
Autophagy-lysosomal pathway deficits contribute to the accumulation of amyloid-β (Aβ), Tau, and lipid droplets in Alzheimer's disease (AD). Capsaicin, a specific agonist of transient receptor potential vanilloid 1 (TRPV1), can improve cognitive function in AD patients, but the detailed mechanism is still unclear. Here, it is revealed that capsaicin ameliorated AD-related pathology by activating peroxisome proliferator-activated receptor alpha (PPARA/PPARα, a key regulator of lipid metabolism) to promote lipid metabolism and reverse autophagy-lysosomal deficits. Molecular mechanism research found that capsaicin significantly activated the PPAR signaling pathway to promote lipid metabolism, with PPARA identified as the key transcription factor. In addition, capsaicin upregulated ATP6V0E1 (V-ATPase V0 complex subunit e1, involved in lysosomal acidification) expression through PPARA, restoring V-ATPase activity. This enhanced lysosomal acidification facilitated lipophagy (autophagic clearance of lipid droplets), while promoting the clearance of Aβ and Tau aggregates via the autophagy-lysosomal pathway. Further, inhibition of ATP6V0E1 and PPARA expression blocked the effect of capsaicin on alleviating AD lipid pathology and cognitive deficits through autophagy-lysosomal flux. Taken together, capsaicin promotes lipid metabolism, reduces lipid deposition, and attenuates AD-related pathologies, while PPARA-ATP6V0E1-V-ATPase signaling mediated autophagy-lysosomal pathway plays a key role in this process.
Keywords: ATP6V0E1; Alzheimer's disease; PPARA; V‐ATPase; autophagy; capsaicin.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Collaborators G. B. D. N., Lancet Neurol. 2019, 18, 459.
-
- Hodson R., Nature 2018, 559, S1. - PubMed
-
- Hamilton L. K., Dufresne M., Joppe S. E., Petryszyn S., Aumont A., Calon F., Barnabe‐Heider F., Furtos A., Parent M., Chaurand P., Fernandes K. J., Cell Stem Cell 2015, 17, 397. - PubMed
-
- Huynh K., Lim W. L. F., Giles C., Jayawardana K. S., Salim A., Mellett N. A., Smith A. A. T., Olshansky G., Drew B. G., Chatterjee P., Martins I., Laws S. M., Bush A. I., Rowe C. C., Villemagne V. L., Ames D., Masters C. L., Arnold M., Nho K., Saykin A. J., Baillie R., Han X., Kaddurah‐Daouk R., Martins R. N., Meikle P. J., Nat. Commun. 2020, 11, 5698. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 82401671/National Natural Science Foundation of China
- 81601121/National Natural Science Foundation of China
- BK20231050/Natural Science Foundation of Jiangsu Province
- BK20211238/Natural Science Foundation of Jiangsu Province
- BK20221201/Natural Science Foundation of Jiangsu Province
- JUSRP123070/Fundamental Research Funds for the Central Universities
- Q202326/Wuxi Municipal Health Commission
- BJ2023040/Wuxi Top Talent Support Program for Young and Middle-aged People of Wuxi Health Committee
- K20231062/Wuxi Science and Technology Development Foundation
- JSTJ-2024-370/Youth Talent Support Project of Jiangsu Province for Science and Technology
- YXXK20240926003/Medical Development Foundation of Jiangnan University
- KYCX25_2779/Postgraduate Research & Practice Innovation Program of Jiangsu Province
LinkOut - more resources
Full Text Sources
Medical
