Constitutive heterochromatin controls nuclear mechanics, morphology, and integrity through H3K9me3 mediated chromocenter compaction
- PMID: 40719377
- PMCID: PMC11988277
- DOI: 10.1080/19491034.2025.2486816
Constitutive heterochromatin controls nuclear mechanics, morphology, and integrity through H3K9me3 mediated chromocenter compaction
Abstract
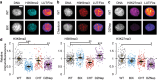
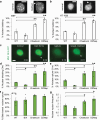
Aberrant nuclear morphology is a hallmark of human disease and causes nuclear dysfunction. Perturbed nuclear mechanics via reduced heterochromatin weakens the nucleus resulting in nuclear blebbing and rupture. While the role of heterochromatin is known, the separate roles of constitutive heterochromatin methylation states remains elusive. Using MEF and HT1080 cells, we isolated the individual contribution of constitutive heterochromatin H3K9 methylation states through histone methyltransferase inhibitors. Inhibition of SUV39H1 via Chaetocin downregulates H3K9 trimethylation (me3), while inhibition of G9a via BIX01294 downregulates H3K9 dimethylation (me2). Overall, the loss of H3K9me3 increased nuclear blebbing and rupture in interphase nuclei due to decreased nuclear rigidity from decompaction of chromocenters. Oppositely, loss of H3K9me2 decreased nuclear blebbing and rupture with increased nuclear rigidity and more compact chromocenters. We show that facultative heterochromatin and HP1α are non-essential for chromocenter compaction. Constitutive heterochromatin provides essential nuclear mechanical support to maintain nuclear shape and integrity through chromocenter compaction.
Keywords: Chromocenter; heterochromatin; nuclear blebbing; nuclear mechanics; nuclear structure.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
SKI regulates rRNA transcription and pericentromeric heterochromatin to ensure centromere integrity and genome stability.Neoplasia. 2025 Sep;67:101204. doi: 10.1016/j.neo.2025.101204. Epub 2025 Jul 2. Neoplasia. 2025. PMID: 40609276 Free PMC article.
-
ASB7 is a negative regulator of H3K9me3 homeostasis.Science. 2025 Jul 17;389(6757):309-316. doi: 10.1126/science.adq7408. Epub 2025 May 29. Science. 2025. PMID: 40440427
-
A mutant ASXL1-BAP1-EHMT complex contributes to heterochromatin dysfunction in clonal hematopoiesis and chronic monomyelocytic leukemia.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2413302121. doi: 10.1073/pnas.2413302121. Epub 2025 Jan 3. Proc Natl Acad Sci U S A. 2025. PMID: 39752521 Free PMC article.
-
Inheritance of H3K9 methylation regulates genome architecture in Drosophila early embryos.EMBO J. 2024 Jul;43(13):2685-2714. doi: 10.1038/s44318-024-00127-z. Epub 2024 Jun 3. EMBO J. 2024. PMID: 38831123 Free PMC article.
-
Nuclear deformability depends on H3K9-methylated heterochromatin anchorage to the nuclear periphery in Caenorhabditis elegans.Genetics. 2025 Jul 9;230(3):iyaf086. doi: 10.1093/genetics/iyaf086. Genetics. 2025. PMID: 40335054 Free PMC article.
Cited by
-
Transcriptional activity generates chromatin motion that drives nuclear blebbing.bioRxiv [Preprint]. 2025 May 21:2025.05.20.655131. doi: 10.1101/2025.05.20.655131. bioRxiv. 2025. PMID: 40475668 Free PMC article. Preprint.
-
Heterogeneity as a feature: unraveling chromatin's role in nuclear mechanics.Nucleus. 2025 Dec;16(1):2545037. doi: 10.1080/19491034.2025.2545037. Epub 2025 Aug 21. Nucleus. 2025. PMID: 40838526 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
