STIM-IP3R crosstalk regulates migration of breast cancer cells
- PMID: 40719699
- PMCID: PMC12302952
- DOI: 10.1083/jcb.202411203
STIM-IP3R crosstalk regulates migration of breast cancer cells
Abstract
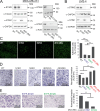
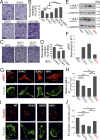
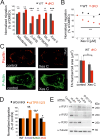
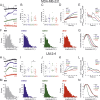
Calcium ions (Ca2+) are crucial second messengers involved in numerous processes including tumorigenesis and cancer cell migration. Previous studies have shown that the endoplasmic reticulum (ER) Ca2+ sensors, stromal interaction molecules STIM1 and STIM2, are key regulators of cancer cell migration. In this study, using breast cancer cells lacking one or both STIM isoforms we show that although STIM proteins are critical regulators of cell migration, they are dispensable for this cellular activity. The mechanism underlying this complex effect involves functional crosstalk between STIM proteins and inositol 1,4,5-trisphosphate receptors (IP3Rs). Our findings indicate that beyond their classical role in store-operated Ca2+ entry, STIM proteins shape the spatial dynamics of IP3R-mediated Ca2+ release. Our results suggest that following ER Ca2+ depletion, the activated STIM proteins shift the pattern of IP3R-mediated Ca2+ release from a localized signal, which promotes cell migration, to a more diffuse signal, which attenuates cell migration.
© 2025 Militsin et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures








References
-
- Ahmad, M., Ong H.L., Saadi H., Son G.-Y., Shokatian Z., Terry L.E., Trebak M., Yule D.I., and Ambudkar I.. 2022. Functional communication between IP3R and STIM2 at subthreshold stimuli is a critical checkpoint for initiation of SOCE. Proc. Natl. Acad. Sci. USA. 119:e2114928118. 10.1073/pnas.2114928118 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

