Genomic insights into bacteriophages: a new frontier in AMR detection and phage therapy
- PMID: 40720171
- PMCID: PMC12302716
- DOI: 10.1093/bfgp/elaf011
Genomic insights into bacteriophages: a new frontier in AMR detection and phage therapy
Abstract
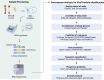
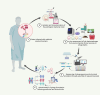
The misuse and overprescription of antibiotics have accelerated the rise of antimicrobial resistance (AMR), rendering many antibiotics ineffective and leading to significant clinical challenges. The conventional treatment methods have become progressively challenging, posing a threat of evolving into an impending silent pandemic. The long track record of bacteriophages combating bacterial infections has renewed hope into the potential therapeutic benefits of bacteriophages. Bacteriophage therapy offers a promising alternative to antibiotics, particularly against multidrug-resistant (MDR) pathogens. This article explores the promise of phages as a potential means to combat superbugs from the perspective of the genomic and transcriptomic landscape of the phages and their bacterial host. Advances in bacteriophage genomics have expedited the detection of new phages and AMR genes, enhancing our understanding of phage-host interactions and enabling the identification of potential treatments for antibiotic-resistant bacteria. At the same time, holo-transcriptomic studies hold potential for discovering disease and context-specific transcriptionally active phages vis-à-vis disease severity. Holo-transcriptomic profiling can be applied to investigate the presence of AMR-bacteria, highlighting COVID-19 and Dengue diseases, in addition to the globally recognized ESKAPE pathogens. By simultaneously capturing phage, bacterial and host transcripts, this approach enables a better comprehension of the bacteriophage dynamics. Moreover, insight into these defence and counter-defence interactions is essential for augmenting the adoption of phage therapy at scale and advancing bacterial control in clinical settings.
Keywords: AMR; Holo-transcriptomics; bacteriophages; disease severity; genomics; phage therapy.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Marine Bacteriophages as Next-Generation Therapeutics: Insights into Antimicrobial Potential and Application.Viruses. 2025 Jul 10;17(7):971. doi: 10.3390/v17070971. Viruses. 2025. PMID: 40733588 Free PMC article. Review.
-
Isolation and characterization of bacteriophages with lytic activity against multidrug-resistant non-typhoidal Salmonella from Nairobi City county, Kenya.BMC Infect Dis. 2025 Jul 24;25(1):940. doi: 10.1186/s12879-025-11325-3. BMC Infect Dis. 2025. PMID: 40707891 Free PMC article.
-
Bacteriophage infection drives loss of β-lactam resistance in methicillin-resistant Staphylococcus aureus.Elife. 2025 Jul 10;13:RP102743. doi: 10.7554/eLife.102743. Elife. 2025. PMID: 40637714 Free PMC article.
-
Bacteriophage therapy- a refurbished age-old potential strategy to treat antibiotic and multidrug resistant bacterial infections in future.Braz J Microbiol. 2024 Sep;55(3):3043-3049. doi: 10.1007/s42770-024-01434-7. Epub 2024 Jul 1. Braz J Microbiol. 2024. PMID: 38951476 Free PMC article.
-
Armed Phages: A New Weapon in the Battle Against Antimicrobial Resistance.Viruses. 2025 Jun 27;17(7):911. doi: 10.3390/v17070911. Viruses. 2025. PMID: 40733529 Free PMC article. Review.
References
-
- World Health Organization . Who. Antimicrobial resistance 2023. Accessed on 19th February 2025.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

