Rare variants in BMAL1 are associated with a neurodevelopmental syndrome
- PMID: 40720646
- PMCID: PMC12337293
- DOI: 10.1073/pnas.2427085122
Rare variants in BMAL1 are associated with a neurodevelopmental syndrome
Abstract
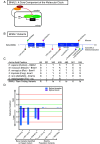
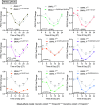
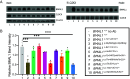
Through international gene-matching efforts, we identified 10 individuals with ultrarare heterozygous variants, including 5 de novo variants, in BMAL1, a core component of the molecular clock. Instead of an isolated circadian phenotype seen with disease-causing variants in other molecular clock genes, all individuals carrying BMAL1 variants surprisingly share a clinical syndrome manifest as developmental delay and autism spectrum disorder, with variably penetrant sleep disturbances, seizures, and marfanoid habitus. Variants were functionally tested in cultured cells using a Per2-promoter driven luciferase reporter and revealed both loss-of-function and gain-of-function changes in circadian rhythms. The tested BMAL1 variants disrupted PER2 mRNA cycling, but did not cause significant shifts in cellular localization or binding with CLOCK. Conserved variants were further tested in Drosophila, which confirmed variant-dependent effects on behavioral rhythms. Remarkably, flies expressing variant cycle, the ortholog of BMAL1, also demonstrated deficits in short- and long-term memory, reminiscent of the highly prevalent developmental delay observed in our cohort. We suggest that ultrarare variants in the BMAL1 core clock gene contribute to a neurodevelopmental disorder.
Keywords: BMAL1; Drosophila; circadian rhythms; developmental delay; neurodevelopmental disorder.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Toh K. L., et al. , An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043 (2001). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

