Genomics of Neotropical biodiversity indicators: Two butterfly radiations with rampant chromosomal rearrangements and hybridization
- PMID: 40720651
- PMCID: PMC12337270
- DOI: 10.1073/pnas.2410939122
Genomics of Neotropical biodiversity indicators: Two butterfly radiations with rampant chromosomal rearrangements and hybridization
Abstract
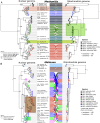
A central question in evolutionary biology is what drives the diversification of lineages. Rapid, recent radiations are ideal systems for this question because they still show key morphological and ecological adaptations associated with speciation. While most research on recent radiations focuses on those occurring in insular environments, less attention has been given to continental radiations with complex species interactions. Here, we study the drivers of continental radiations of Melinaea and Mechanitis butterflies (Nymphalidae: Ithomiini), which have rapidly radiated in the continental Neotropics. They are classical models for Amazonian biogeography and color pattern mimicry and have been proposed as biodiversity indicators. We generated reference genomes for five species of each genus and whole-genome resequencing data of most species and subspecies covering a wide geographic range to assess phylogeographic relationships, hybridization patterns, and chromosomal rearrangements. Our data help resolve the classification of these taxonomically challenging butterflies and reveal very high diversification rates. We find rampant evidence of historical hybridization and putative hybrid species in both radiations, which may have facilitated their rapid diversification by enriching the genetic diversity. Moreover, we identified dozens of chromosomal fusions and fissions between congeneric species that have likely expedited reproductive isolation. We conclude that interactions between geography, hybridization and chromosomal rearrangements have contributed to these rapid radiations in the highly diverse Neotropical region. We hypothesize that rapid radiations may be spurred if repeated periods of geographic isolation are combined with lineage-specific rapid accumulation of incompatibilities, followed by secondary contact with some gene exchange.
Keywords: Neotropics; biodiversity; butterflies; genomics; speciation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Losos J. B., Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 175, 623–639 (2010). - PubMed
-
- Gittenberger E., Radiation and adaptation, evolutionary biology and semantics. Organ. Divers. Evol. 4, 135–136 (2004).
-
- Czekanski-Moir J. E., Rundell R. J., The ecology of nonecological speciation and nonadaptive radiations. Trends Ecol. Evol. 34, 400–415 (2019). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

