Inhibition of HDAC6 alters fumarate hydratase activity and mitochondrial structure
- PMID: 40721560
- PMCID: PMC12304134
- DOI: 10.1038/s41467-025-61897-6
Inhibition of HDAC6 alters fumarate hydratase activity and mitochondrial structure
Abstract
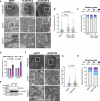
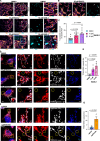
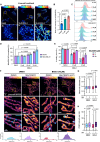
Fumarate hydratase (FH), a key node of mitochondrial metabolism, is also a tumour suppressor. Despite its prominent roles in tumourigenesis and inflammation, its regulation remains poorly understood. Herein, we show that histone deacetylase 6 (HDAC6) regulates FH activity. In triple-negative breast cancer cells, HDAC6 inhibition or knockdown results in alterations to mitochondrial cristae structure, as detected by live-cell super-resolution STED nanoscopy and electron microscopy, along with the release of mitochondrial DNA. Mass-spectrometry immunoprecipitation reveals multiple mitochondrial HDAC6-interactors, with FH emerging as a top hit. Super-resolution 3D-STORM shows HDAC6 interactions with FH in mitochondrial networks, which increases after perturbation of HDAC6 activity with BAS-2. Treatment with BAS-2 leads to fumarate accumulation by 13C glucose labelling, along with downstream succination of proteins and cell death. Together, these results identify HDAC6 inhibition as a regulator of endogenous FH activity in tumour cells, and highlight it as a promising candidate for indirectly targeting tumour metabolism.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Gonçalves, A. C. et al. Impact of cancer metabolism on therapy resistance - Clinical implications. Drug Resist Updat59, 100797 (2021). - PubMed
-
- Gallinari, P., Di Marco, S., Jones, P., Pallaoro, M. & Steinkühler, C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res17, 195–211 (2007). - PubMed
-
- Rodrigues, D. A., Roe, A., Griffith, D. & Chonghaile, T. N. Advances in the design and development of PROTAC-mediated HDAC degradation. Curr. Top. Med Chem.22, 408–424 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

