CD142-positive synovial fibroblasts drive meniscus destruction in rheumatoid arthritis
- PMID: 40721606
- PMCID: PMC12304273
- DOI: 10.1038/s41467-025-61842-7
CD142-positive synovial fibroblasts drive meniscus destruction in rheumatoid arthritis
Abstract
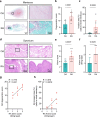
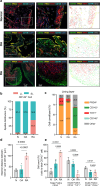
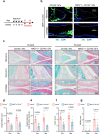
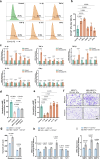
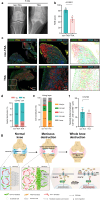
Previous evidence suggest bone and cartilage damage is the main pathogenesis of rheumatoid arthritis joint destruction. However, the role of meniscus usually has not been thoroughly explored. Here, we identify CD142+ synovial fibroblasts as a subset located at sublining layer in normal and osteoarthritis synovium, which is increased and distributed at lining layer in rheumatoid arthritis synovium. Injection of CD142+ fibroblasts into DBA/1 male mice's knee destructs meniscus but has slight effect on cartilage. ABCC4 is highly expressed in CD142+ fibroblasts, whose blockage by MK571 attenuates CD142+ fibroblasts-induced meniscus destruction through cAMP/PKA signaling. Long-term follow-up of rheumatoid arthritis cohort indicates that enriched CD142+ fibroblasts at lining layer are a risk factor for severe knee joint destruction and eventually undergo total knee arthroplasty. Our results demonstrate CD142+ fibroblasts as an indicator to assess prognosis and a therapeutic target to inhibit meniscal destruction, thereby alleviating rheumatoid arthritis knee joint damage.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

