EWS::FLI1-DHX9 interaction promotes Ewing sarcoma sensitivity to DNA topoisomerase 1 poisons by altering R-loop metabolism
- PMID: 40721661
- PMCID: PMC12436182
- DOI: 10.1038/s41388-025-03496-9
EWS::FLI1-DHX9 interaction promotes Ewing sarcoma sensitivity to DNA topoisomerase 1 poisons by altering R-loop metabolism
Abstract
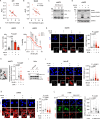
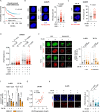
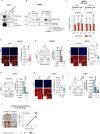
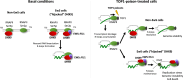
Drug resistance is an ill-defined cause of dismal outcomes in cancer. Ewing sarcoma (EwS), a pediatric cancer characterized by high therapy failure rates, is driven by a single oncogenic event generating EWSR1::ETS gene fusions (primarily EWSR1::FLI1) in a silent genomic background. This provides a straightforward model to study the impact of gene fusions on drug responses. Here, we describe a novel mechanism of sensitivity to DNA topoisomerase 1 poisons in EwS. We discovered that EWS::FLI1 prevents the resolution of R-loops induced by these drugs via sequestering DHX9 helicase, ultimately resulting in R-loop accumulation, replication stress, and genome instability. In turn, excessive DHX9 or reduced EWS::FLI1 levels render EwS cells resistant to the active metabolite of irinotecan (SN-38) independent of proliferation and global transcription rates. This resistance helps explain how elevated DHX9 levels predict worse clinical outcomes. Overall, our research demonstrates the impact of a dominant mutation on cancer drug sensitivity, highlighting its significant clinical implications.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All methods were performed following the relevant guidelines and regulations, including the Declaration of Helsinki for research involving human participants and Directive 2010/63/EU for the protection of animals used for scientific purposes. The study involving human participants was approved by the Comité Coordinador de Ética de la Investigación Biomédica de Andalucía, which issued a favorable opinion on April 10, 2024, for the project (Protocol version 1, dated 04/06/2023; IC version 7, dated 18/05/2021; internal code SICEIA-2024-000125; application code S2400018). Written informed consent was obtained from all participants before their inclusion in the study. The study also included research involving live vertebrates (mice, Mus musculus). It was authorized by the Dirección General de la Producción Agrícola y Ganadera of the Consejería de Agricultura, Pesca, Agua y Desarrollo Rural of the Junta de Andalucía. The corresponding ethics approval (reference number: 05/07/2023/58) was granted on July 6, 2023, for the project, classified as a Type II project involving mild to moderate procedures. The protocol was positively evaluated by the local Animal Ethics Committee and by the authorized body CEI de los Hospitales Universitarios Virgen Macarena - Virgen del Rocío de Sevilla. No identifiable images of human participants are included in this manuscript.
Figures


References
-
- Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers. 2018;4:5. - PubMed
-
- Esiashvili N, Goodman M, Marcus RB Jr. Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J Pediatr Hematol Oncol. 2008;30:425–30. - PubMed
-
- Gaspar N, Hawkins DS, Dirksen U, Lewis IJ, Ferrari S, Le Deley MC, et al. Ewing sarcoma: current management and future approaches through collaboration. J Clin Oncol. 2015;33:3036–46. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

