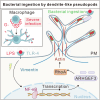
Macrophages form dendrite-like pseudopods to enhance bacterial ingestion
- PMID: 40721684
- PMCID: PMC12402336
- DOI: 10.1038/s44318-025-00515-z
Macrophages form dendrite-like pseudopods to enhance bacterial ingestion
Abstract
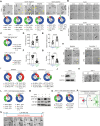
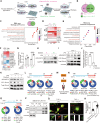
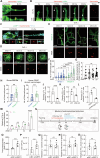
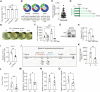
Macrophages are critical innate immune cells that exhibit remarkable adaptability during pathogen infections. However, the relationship between their morphological plasticity and physiological functions remains largely elusive. Here, we discovered an unprecedented paradigm of macrophage adaptation within a few hours upon severe Gram-negative bacterial infections, characterized by the formation of dendrite-like pseudopods (DLPs). Using in vitro, microfluidic, and in vivo infection models, we demonstrate that these pseudopods enhance bacterial uptake by expanding the macrophage searching radius, thereby bolstering host defense. Mechanistically, Toll-like receptor 4 (TLR4) activation by Gram-negative bacterial lipopolysaccharide (LPS) upregulates the expression of macrophage-specific RhoGEF and ARHGEF3 in an NF-κB-dependent manner. ARHGEF3 localizes to dendrite-like pseudopods and enhances RhoA activity. Consequently, periodic cycles of actin assembly and disassembly propel the elongation of pseudopods, whereas vimentin intermediate filaments stabilize them. Importantly, infusion of DLP-equipped macrophages into Salmonella-infected mice reduced bacterial burden and infection severity. Together, our findings underscore how the dynamic response of macrophages to massive infections can augment immune defense against pathogenic bacteria.
Keywords: Actin Filaments; Dendrite-like Pseudopods; Macrophage; Pathogen Ingestion; Vimentin Intermediate Filaments.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures





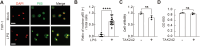

References
-
- Anes E (2017) Acting on actin during bacterial infection. In: Jimenez-Lopez JC (ed.) Cytoskeleton—structure, dynamics, function and disease. IntechOpen, Rijeka. pp 27–44
-
- Arthur WT, Ellerbroek SM, Der CJ, Burridge K, Wennerberg K (2002) XPLN, a guanine nucleotide exchange factor for RhoA and RhoB, but not RhoC. J Biol Chem 277:42964–42972 - PubMed
-
- Binley KE, Ng WS, Tribble JR, Song B, Morgan JE (2014) Sholl analysis: a quantitative comparison of semi-automated methods. J Neurosci methods 225:65–70 - PubMed
MeSH terms
Substances
Grants and funding
- 32222022/MOST | National Natural Science Foundation of China (NSFC)
- 92354301/MOST | National Natural Science Foundation of China (NSFC)
- 23ZR1470900/STCSM | Natural Science Foundation of Shanghai Municipality ()
- GZNL2023A03004/R&D Program of Guangzhou National Laboratory
- 2024YFC2310003/MOST | National Key Research and Development Program of China (NKPs)
LinkOut - more resources
Full Text Sources
Medical

