Prediction of Parkinson Disease Using Long-Term, Short-Term Acoustic Features Based on Machine Learning
- PMID: 40722330
- PMCID: PMC12294038
- DOI: 10.3390/brainsci15070739
Prediction of Parkinson Disease Using Long-Term, Short-Term Acoustic Features Based on Machine Learning
Abstract
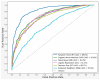
Background: Parkinson's disease (PD) is the second most common neurodegenerative disorder after Alzheimer's disease, affecting countless individuals worldwide. PD is characterized by the onset of a marked motor symptomatology in association with several non-motor manifestations. The clinical phase of the disease is usually preceded by a long prodromal phase, devoid of overt motor symptomatology but often showing some conditions such as sleep disturbance, constipation, anosmia, and phonatory changes. To date, speech analysis appears to be a promising digital biomarker to anticipate even 10 years before the onset of clinical PD, as well serving as a useful prognostic tool for patient follow-up. That is why, the voice can be nominated as the non-invasive method to detect PD from healthy subjects (HS). Methods: Our study was based on cross-sectional study to analysis voice impairment. A dataset comprising 81 voice samples (41 from healthy individuals and 40 from PD patients) was utilized to train and evaluate common machine learning (ML) models using various types of features, including long-term (jitter, shimmer, and cepstral peak prominence (CPP)), short-term features (Mel-frequency cepstral coefficient (MFCC)), and non-standard measurements (pitch period entropy (PPE) and recurrence period density entropy (RPDE)). The study adopted multiple machine learning (ML) algorithms, including random forest (RF), K-nearest neighbors (KNN), decision tree (DT), naïve Bayes (NB), support vector machines (SVM), and logistic regression (LR). Cross-validation technique was applied to ensure the reliability of performance metrics on train and test subsets. These metrics (accuracy, recall, and precision), help determine the most effective models for distinguishing PD from healthy subjects. Result: Among all the algorithms used in this research, random forest (RF) was the best-performing model, achieving an accuracy of 82.72% with a ROC-AUC score of 89.65%. Although other models, such as support vector machine (SVM), could be considered with an accuracy of 75.29% and a ROC-AUC score of 82.63%, RF was by far the best one when evaluated across all metrics. The K-nearest neighbor (KNN) and decision tree (DT) performed the worst. Notably, by combining a comprehensive set of long-term, short-term, and non-standard acoustic features, unlike previous studies that typically focused on only a subset, our study achieved higher predictive performance, offering a more robust model for early PD detection. Conclusions: This study highlights the potential of combining advanced acoustic analysis with ML algorithms to develop non-invasive and reliable tools for early PD detection, offering substantial benefits for the healthcare sector.
Keywords: Parkinson’s disease; machine learning; mel-frequency cepstral coefficient; vocal features.
Conflict of interest statement
Authors Andrea Buccoliero and Marcello Dorian Donzella were employed by the company GPI S.p.A. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Chaudhuri K.R., Azulay J.P., Odin P., Lindvall S., Domingos J., Alobaidi A., Kandukuri P.L., Chaudhari V.S., Parra J.C., Yamazaki T., et al. Economic Burden of Parkinson’s Disease: A Multinational, Real-World, Cost-of-Illness Study. Drugs Real World Outcomes. 2024;11:1–11. doi: 10.1007/s40801-023-00410-1. - DOI - PMC - PubMed
-
- Mallamaci R., Musarò D., Greco M., Caponio A., Castellani S., Munir A., Guerra L., Damato M., Fracchiolla G., Coppola C., et al. Dopamine- and Grape-Seed-Extract-Loaded Solid Lipid Nanoparticles: Interaction Studies between Particles and Differentiated SH-SY5Y Neuronal Cell Model of Parkinson’s Disease. Molecules. 2024;29:1774. doi: 10.3390/molecules29081774. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources