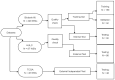
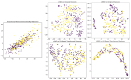
OKEN: A Supervised Evolutionary Optimizable Dimensionality Reduction Framework for Whole Slide Image Classification
- PMID: 40722425
- PMCID: PMC12292405
- DOI: 10.3390/bioengineering12070733
OKEN: A Supervised Evolutionary Optimizable Dimensionality Reduction Framework for Whole Slide Image Classification
Abstract
Classification of lung cancer subtypes is a critical clinical step; however, relying solely on H&E-stained histopathology images can pose challenges, and additional immunohistochemical analysis is sometimes required for definitive subtyping. Digital pathology facilitates the use of artificial intelligence for automatic classification of digital tissue slides. Automatic classification of Whole Slide Images (WSIs) typically involves extracting features from patches obtained from them. The aim of this study was to develop a WSI classification framework utilizing an optimizable kernel to encode features from each patch from a WSI into a desirable and adjustable latent space using an evolutionary algorithm. The encoded data can then be used for classification and segmentation while being computationally more efficient. Our proposed framework is compared with a state-of-the-art model, Vim4Path, on an internal and external dataset of lung cancer WSIs. The proposed model outperforms Vim-S16 in accuracy and F1 score at both ×2.5 and ×10 magnification levels on the internal test set, with the highest accuracy (0.833) and F1 score (0.721) at ×2.5. On the external test set, Vim-S16 at ×10 achieves the highest accuracy (0.732), whereas OKEN-DenseNet121 at ×2.5 has the best F1 score (0.772). In future work, finding a dynamic way to tune the output dimensions of the evolutionary algorithm would be of value.
Keywords: deep learning; digital pathology; dimensionality reduction; evolutionary algorithm; lung cancer.
Conflict of interest statement
Author André Pedersen was employed by Sopra Steria. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Strauss G.M., Jemal A., McKenna M.B., Strauss J.A., Cummings K. B4-06: Lung Cancer Survival in Relation to Histologic Subtype: An Analysis based upon Surveillance Epidemiology and End Results (SEER) Data. J. Thorac. Oncol. 2007;2:S345–S346. doi: 10.1097/01.JTO.0000283165.65986.59. - DOI
-
- Yun J.K., Kwon Y., Kim J., Lee G.D., Choi S., Kim H.R., Kim Y.H., Kim D.K., Park S.I. Clinical impact of histologic type on survival and recurrence in patients with surgically resected stage II and III non-small cell lung cancer. Lung Cancer. 2023;176:24–30. doi: 10.1016/j.lungcan.2022.12.008. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

