DIRAS1 Drives Oxaliplatin Resistance in Colorectal Cancer via PHB1-Mediated Mitochondrial Homeostasis
- PMID: 40723377
- PMCID: PMC12292402
- DOI: 10.3390/biology14070819
DIRAS1 Drives Oxaliplatin Resistance in Colorectal Cancer via PHB1-Mediated Mitochondrial Homeostasis
Abstract
Background: Colorectal cancer (CRC) is a prevalent global malignancy with particularly challenging treatment outcomes in advanced stages. Oxaliplatin (OXA) is a frontline chemotherapeutic agent for CRC. However, 15% to 50% of stage III patients experience recurrence due to drug resistance. Elucidating the molecular mechanisms underlying OXA resistance is, therefore, crucial for improving CRC prognosis. The role of DIRAS1, a RAS superfamily member with reported tumor-suppressive functions in various cancers, remains poorly defined in CRC.
Methods: The effects of DIRAS1 on CRC cell proliferation and migration were evaluated using MTT, wound healing, and colony formation assays. Stable cell lines with knockdown or overexpression of DIRAS1 and PHB1 were established via plasmid and lentiviral systems. Drug sensitivity to OXA was assessed through cytotoxicity assays and IC50 determination. Clinical relevance was validated through immunohistochemical analysis of CRC tissue samples. Transcriptomic sequencing was performed to explore downstream regulatory mechanisms.
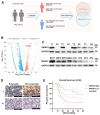
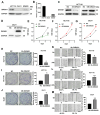
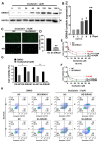
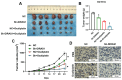
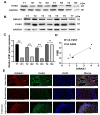
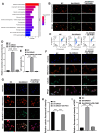
Results: DIRAS1 expression was positively correlated with OXA resistance and was significantly upregulated following prolonged chemotherapy exposure. Silencing DIRAS1 reduced the IC50 of OXA in vitro and increased tumor sensitivity to OXA in vivo. Transcriptome analysis identified PHB1 as a downstream effector of DIRAS1. Functional studies revealed that PHB1 contributes to chemoresistance by maintaining mitochondrial stability.
Conclusions: This study identifies DIRAS1 as a key contributor to OXA resistance in CRC by modulating PHB1 expression and mitochondrial function. Targeting the DIRAS1-PHB1 axis may offer a novel therapeutic strategy to overcome chemoresistance in CRC.
Keywords: DIRAS1; PHB1; colorectal cancer; mitochondrial function; oxaliplatin resistance; targeted therapy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Phosphorylation of KIAA1429 promotes oxaliplatin resistance through activating the FZD7-Wnt signaling in BRAFV600E-mutated colorectal cancer.J Exp Clin Cancer Res. 2025 Jul 3;44(1):187. doi: 10.1186/s13046-025-03449-w. J Exp Clin Cancer Res. 2025. PMID: 40611274 Free PMC article.
-
Prohibitin 1 inhibits cell proliferation and induces apoptosis via the p53-mediated mitochondrial pathway in vitro.World J Gastrointest Oncol. 2024 Feb 15;16(2):398-413. doi: 10.4251/wjgo.v16.i2.398. World J Gastrointest Oncol. 2024. PMID: 38425403 Free PMC article.
-
USP6NL knockdown suppresses colorectal cancer progression by inducing CASP9-Mediated apoptosis and disrupting FOXC2/SNAI1-Driven EMT and angiogenesis.Funct Integr Genomics. 2025 Jul 11;25(1):153. doi: 10.1007/s10142-025-01663-5. Funct Integr Genomics. 2025. PMID: 40643716
-
The use of irinotecan, oxaliplatin and raltitrexed for the treatment of advanced colorectal cancer: systematic review and economic evaluation.Health Technol Assess. 2008 May;12(15):iii-ix, xi-162. doi: 10.3310/hta12150. Health Technol Assess. 2008. PMID: 18462574
-
Contrast-enhanced ultrasound using SonoVue® (sulphur hexafluoride microbubbles) compared with contrast-enhanced computed tomography and contrast-enhanced magnetic resonance imaging for the characterisation of focal liver lesions and detection of liver metastases: a systematic review and cost-effectiveness analysis.Health Technol Assess. 2013 Apr;17(16):1-243. doi: 10.3310/hta17160. Health Technol Assess. 2013. PMID: 23611316 Free PMC article.
References
-
- Yang L., Zou S., Shu C., Song Y., Sun Y.K., Zhang W., Zhou A., Yuan X., Yang Y., Hu S. CYP2A6 Polymorphisms Associate with Outcomes of S-1 Plus Oxaliplatin Chemotherapy in Chinese Gastric Cancer Patients. Genom. Proteom. Bioinform. 2017;15:255–262. doi: 10.1016/j.gpb.2016.11.004. - DOI - PMC - PubMed
-
- Wang X., Xu Z., Wang J., Wu C., Zhang L., Qian C., Luo Y., Gu Y., Wong W.T., Xiang D. A Mitochondria-Targeted Biomimetic Nanomedicine Capable of Reversing Drug Resistance in Colorectal Cancer Through Mitochondrial Dysfunction. Adv. Sci. 2025;12:e2410630. doi: 10.1002/advs.202410630. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

