Differentiation of mtDNA Methylation in Tissues of Ridgetail White Prawn, Exopalaemon carinicauda
- PMID: 40723500
- PMCID: PMC12291886
- DOI: 10.3390/ani15142037
Differentiation of mtDNA Methylation in Tissues of Ridgetail White Prawn, Exopalaemon carinicauda
Abstract
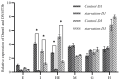
In a previous study, we found that mitochondrial DNA methylation occurred in the muscle tissue of Exopalaemon carinicauda under starvation stress. To explore whether this phenomenon also existed in other tissues, we used the bisulfite method (BSP) to detect the methylation of the mitochondrial genome in the intestinal tissues, hepatopancreas, gills, eye stalks, muscles, heart, and other tissues before and after starvation. In situ hybridization and qPCR techniques were used to analyze the expression of DNMT1 and DNMT3b involved in methylation regulation in different tissues. The results showed that the methylation rate was highest in intestinal tissue, followed by hepatopancreas, gills, heart, muscle, and eye stalk. Significantly different expression levels of DNMT1 and DNMT3b were found in the intestine and hepatopancreas with a higher expression pre-starvation and a lower expression post starvation. The expression levels of DNMT1 and DNMT3b in heart and muscle increased after starvation. The expression levels of DNMT1 and DNMT3b in the eye stalk were low and decreased significantly after starvation. The in situ hybridization of DNMT1 and DNMT3b further verified the results: the mRNA signal in intestinal and hepatopancreatic tissues of the starvation group was significantly weaker than that of the control group. No significant difference in mRNA signal intensity was found in the gill, muscle, and heart tissues of the starvation group compared with the control group. The mRNA signal in the eye stalk tissue of the starvation group was weaker than that of the control group. This study is the first to confirm different levels of mtDNA methylation in different tissues of E. carinicauda, which may be closely related to their biological functions.
Keywords: BSP sequencing; DNA methylation; DNA methyltransferases; Exopalaemon carinicauda; Mitochondria genome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







Similar articles
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Sertindole for schizophrenia.Cochrane Database Syst Rev. 2005 Jul 20;2005(3):CD001715. doi: 10.1002/14651858.CD001715.pub2. Cochrane Database Syst Rev. 2005. PMID: 16034864 Free PMC article.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources

